Figures & data
Table 1. Primers used for cloning of Native, Lipid-modified (Sec & Tat) and Sec mutant recombinant constructs.
Figure 1. (A) Schematic representation of the cloning strategy employed to generate constructs, pRSETB-ICP11, pRSETB-VP28, pRSETB-VP281, pG-LMICP11, pG-LMVP28, pG-LMVP281, pG-TLICP11, pG-TLVP28 and pG-TLVP281. (i) Vector map of the prokaryotic expression vector pRSETB in which the genes of native ICP11, VP28 and VP281 were cloned separately between NdeI and PstI sites. (ii) and (iii) Maps of modified prokaryotic expression vector pRSETB showing the fusion of the coding sequence of lipoprotein signal peptide for Sec (pG-LM) and Tat (pG-TL) respectively upstream of target protein’s coding sequence in the MCS between BamHI and PstI sites. (B) The coding sequences of Sec and Tat signal peptide, Lipobox and several residues after invariant lipid-modifiable Cysteine. All the three enlarged versions of the vector maps are enclosed in the Supplementary information (available online only). (C) Agarose gel electrophoresis of the PCR-amplified products of ICP11, VP28 and VP281: (i) PCR-amplified gene products with 5′ NdeI and 3′ PstI sites meant for mature sequence of native proteins, and (ii) PCR-amplified gene products with 5′ BamHI and 3′PstI sites meant for lipid-modification.
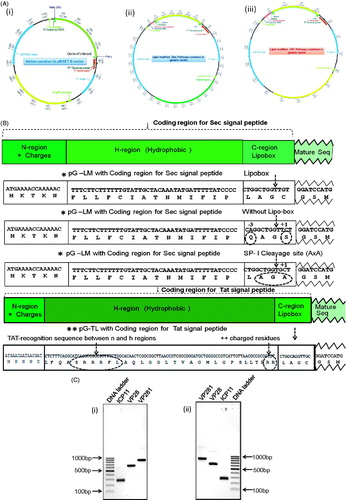
Figure 2. (A) SDS-PAGE profile showing the expression of native and lipid-modified (i) ICP11, and (ii) VP28 and VP281. The corresponding protein migration on the gel is indicated by white arrows. (B) Western blot analysis using specific antibodies showing the expression of the native and lipid-modified ICP11, VP28 and VP281 proteins. (C) Autoradiography of the [9, 10-3H] palmitic acid labelled proteins 1, 2 and 3 correspond to ICP11, VP28 and VP281 respectively, confirming the enzymatic post-translational lipid-modification. Radiolabelling was observed only in the case of Tat-ICP11, Tat-VP28, Tat-VP281 and Sec-ICP11.
![Figure 2. (A) SDS-PAGE profile showing the expression of native and lipid-modified (i) ICP11, and (ii) VP28 and VP281. The corresponding protein migration on the gel is indicated by white arrows. (B) Western blot analysis using specific antibodies showing the expression of the native and lipid-modified ICP11, VP28 and VP281 proteins. (C) Autoradiography of the [9, 10-3H] palmitic acid labelled proteins 1, 2 and 3 correspond to ICP11, VP28 and VP281 respectively, confirming the enzymatic post-translational lipid-modification. Radiolabelling was observed only in the case of Tat-ICP11, Tat-VP28, Tat-VP281 and Sec-ICP11.](/cms/asset/c4932f6b-75df-4394-819a-08f05b9331bb/imbc_a_943819_f0002_b.jpg)
Figure 3. (A) Agarose gel electrophoresis confirming the transcription of Sec-VP28 and Sec-VP281 genes (lanes 3 and 5 respectively). In this experiment total RNA was extracted from the Sec-VP28 and Sec-VP281 recombinants in E. coli BL21 (DE3) host and converted into cDNA by RT-PCR, and was further used as a template for PCR using the corresponding primers. (B) Western-Immunoblot analysis of the induced whole cell lysate of E. coli BL21 (DE3) from the same batch of culture used for the above RNA preparation showing that in the case of Sec-mediated lipid-modification of VP28 and VP281 (lanes 2 and 6), the proteins were not even expressed. (C) Agarose gel electrophoresis confirming the transcription of Sec signal peptide without Lipobox and Sec signal peptide with signal peptidase-I cleavage site (AxA) of VP28 and VP281 recombinants constructs. The transcripts of VP28 and VP281 were loaded into lanes 2 & 3 and 6 & 7, lanes 1 and 5 were PCR reaction positive controls of VP28 and VP281 respectively and lane 4 was 100 bp DNA ladder. (D) SDS-PAGE profile showing the expression of VP28 and VP281 with Sec signal peptide without Lipobox and Sec signal peptide with signal peptidase-I cleavage site (AxA). The corresponding protein expression on the gel is indicated by arrows. In the case of VP28 the targeted protein expression was lower the detectable limit by vision. (E) Western-immunoblot analysis confirm the induced, uninduced whole cell lysate of E. coli BL21(DE3) and periplasmic fraction of VP28 and VP281 of targeted proteins. The corresponding protein expression was not detected in uninduced whole cell lysate of E. coli BL21(DE3) and the corresponding proteins were secreted in the periplasmic space.
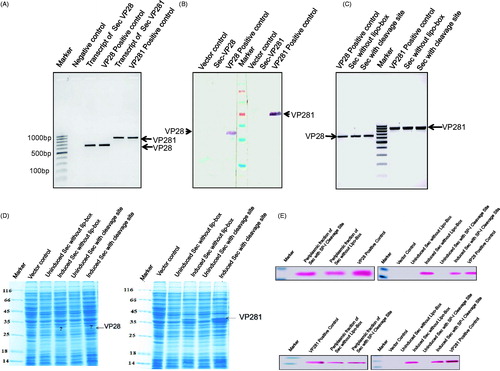
Table 2. Comprehensive analysis of protein expression, its translocation and stability of mRNA.
Figure 4. (A) SDS-PAGE profile of 100 mg of all fractions showing the expression and sub-cellular localization (see arrows) of (i) Sec-ICP11, (ii) Tat-ICP11, (iii) Tat-VP28, and (iv) Tat-VP281 cloned with (for lipid modification) and without (native) the corresponding signal peptide. All the native proteins were expressed in the cytosol (see second lane in all the pictures). The lipid-modified proteins were localized to the inner membrane (see 8th lane in all the pictures). (B) Western-immunoblot analysis to ascertain the sub-cellular localization of the respective proteins. Monoclonal anti-VP28 antibody was used for probing VP28 proteins and monoclonal anti-his antibody for the rest of the proteins. Cyto, cytoplasm; IM, inner membrane; OM, outer membrane; M, marker.
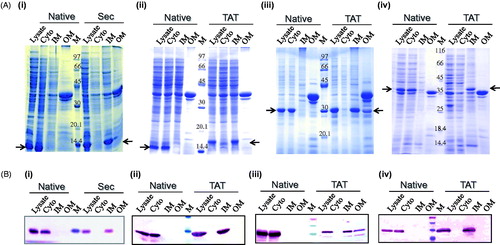
Figure 5. Bar graph from the triplicate experiment representing the extent of solubilization of lipid-modified (i) Tat-ICP11, (ii) Tat-VP28, and (iii) Tat-VP281 in different solubilizing agents: β-Octyl glucoside (OG), Sodium lauoryl sarcocine (SLS) and Urea. The black bars represent the percentage of soluble fraction and the grey bars represent the percentage of insoluble fraction.
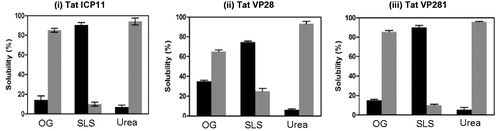
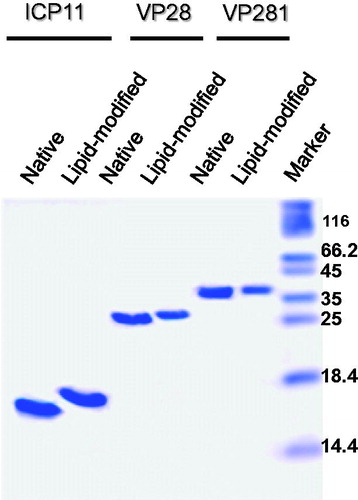
Figure 6. Tricine-SDS-PAGE profile showing the migration difference between purified native (lanes 1,3,5) and lipid-modified (lanes 2,4,6) ICP11, VP28 and VP281. Apparently homogenous preparations in the soluble state can be deduced from the analysis of 5 µg (VP28 and VP281) and 10 µg (ICP11) proteins loaded.