Figures & data
Figure 1. Structures of detergents and of a protein-micelle complex. (A) Chemical structures of the detergents sodium dodecyl sulphate (SDS), lauryldimethylamine-oxide (LDAO), n-octyl-β-D-glucoside (β-OG), n-dodecyl-β-D-maltoside (DDM) and fos-cholines -10, -11, -12 and -14. The pink areas indicate the hydrophilic head groups. (B) Illustration of a protein-detergent micelle complex: Molecular dynamics simulation of the outer membrane β-barrel protein OmpA from Escherichia coli in a dodecylphosphocholine micelle. The picture of the OmpA-micelle complex was reproduced with permission from Bond and Sansom (Citation2003), which was originally published in JMB (Bond PJ, Sansom MS. 2003. Membrane protein dynamics versus environment: Simulations of OmpA in a micelle and in a bilayer. J Mol Biol 329:1035–1053), copyright by Elsevier Science Ltd 2003. This Figure is reproduced in colour in the online version of Molecular Membrane Biology.
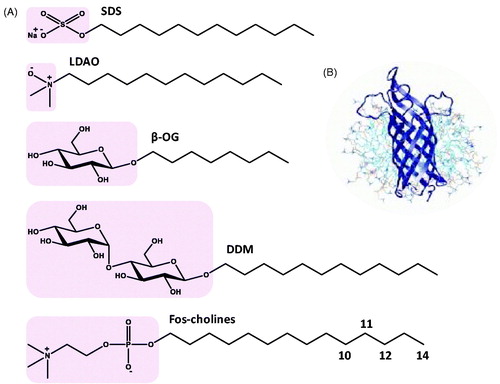
Table I. Properties of detergents. These properties are for the undeuterated compounds and were obtained from catalogues of the product suppliers: Anatrace, Cambridge Isotope Laboratories, Cortecnet, Generon, Sigma-Aldrich.
Figure 2. Synthesis of d25-SDS and its use in solubilizing helix A from brome mosaic virus protein 1a and the C-terminal region of the Frizzled receptor 1. (A) Synthesis of d25-SDS from d25-n-dodecanol. The grey area shows the region of deuteration. (B) (i) [15N-1H]HSQC spectrum with assignments of brome mosaic virus 1a helix A bound to 100 mM d25-SDS micelles and (ii) ensemble of 20 structures (backbone atoms only) determined for helix A bound to an SDS micelle where white = helix and grey = coil. This picture was modified from Liu et al. (Citation2009), which was originally published in PLoS Pathog (Liu L, Westler WM, den Boon JA, Wang X, Diaz A, Steinberg HA, Ahlquist P. 2009. An amphipathic alpha-helix controls multiple roles of brome mosaic virus protein 1a in RNA replication complex assembly and function. PLoS Pathog 5:e1000351), copyright by Liu et al., Citation2009. (C) (i) [15N-1H]HSQC spectrum with assignments of the C-terminal region of the Frizzled receptor 1 (residues 623-647) in d25-SDS micelles, (ii) NOESY spectrum with NOE interactions labelled and (iii) structure determined from 104 NOE restraints. This picture was modified from Gayen et al. (Citation2013) which was originally published in Molecules (Gayen S, Li Q, Kim YM, Kang C. 2013. Structure of the C-terminal region of the Frizzled receptor 1 in detergent micelles. Molecules 18:8579–8590), copyright by Gayen et al., Citation2013. This Figure is reproduced in colour in the online version of Molecular Membrane Biology.
![Figure 2. Synthesis of d25-SDS and its use in solubilizing helix A from brome mosaic virus protein 1a and the C-terminal region of the Frizzled receptor 1. (A) Synthesis of d25-SDS from d25-n-dodecanol. The grey area shows the region of deuteration. (B) (i) [15N-1H]HSQC spectrum with assignments of brome mosaic virus 1a helix A bound to 100 mM d25-SDS micelles and (ii) ensemble of 20 structures (backbone atoms only) determined for helix A bound to an SDS micelle where white = helix and grey = coil. This picture was modified from Liu et al. (Citation2009), which was originally published in PLoS Pathog (Liu L, Westler WM, den Boon JA, Wang X, Diaz A, Steinberg HA, Ahlquist P. 2009. An amphipathic alpha-helix controls multiple roles of brome mosaic virus protein 1a in RNA replication complex assembly and function. PLoS Pathog 5:e1000351), copyright by Liu et al., Citation2009. (C) (i) [15N-1H]HSQC spectrum with assignments of the C-terminal region of the Frizzled receptor 1 (residues 623-647) in d25-SDS micelles, (ii) NOESY spectrum with NOE interactions labelled and (iii) structure determined from 104 NOE restraints. This picture was modified from Gayen et al. (Citation2013) which was originally published in Molecules (Gayen S, Li Q, Kim YM, Kang C. 2013. Structure of the C-terminal region of the Frizzled receptor 1 in detergent micelles. Molecules 18:8579–8590), copyright by Gayen et al., Citation2013. This Figure is reproduced in colour in the online version of Molecular Membrane Biology.](/cms/asset/b5b5f0a2-349a-4c4e-a00d-cb4e16e5c39a/imbc_a_1125536_f0002_c.jpg)
Figure 3. Synthesis of d31-LDAO and its use in solubilizing the human voltage-dependent anion channel (VDAC-1) and use of tail-deuterated decyl-N,N′-dimethyl amine oxide in solubilizing Escherichia coli outer membrane protein OmpF. (A) Synthesis of d31-LDAO from d23-dodecanoic acid. The grey areas show the regions of deuteration. (B) (i) [15N-1H]-TROSY spectrum of [U-2H,15N]VDAC-1 in d31-LDAO micelles, (ii) [13C-1H]HMQC spectrum of [U-2H,13C,15N; 1Hδ-IL; 1Hγ-V]VDAC-1 in d31-LDAO micelles highlighting the spectral regions for Ile residues (red box, 11/11 assigned) and Leu plus Val residues (blue box, 8/12 Val and 17/28 Leu assigned), (iii) structure of human VDAC-1 shown as a side view (top) and from above (bottom) with the N-terminus in blue and C-terminus in red, which were drawn using PDB file 2K4T and PDB Protein Workshop 3.9 (Moreland et al., Citation2005). Pictures of the spectra were reproduced with permission from Hiller et al. (Citation2008), which were originally published in Science (Hiller S, Garces RG, Malia TJ, Orekhov VY, Colombini M, Wagner G. 2008. Solution structure of the integral human membrane protein VDAC-1 in detergent micelles. Science 321:1206–1210, copyright by American Association for the Advancement of Science 2008. (C) (i) Crystal structure of the E. coli outer membrane protein OmpF in tetragonal crystal form as an above view with the N-terminus in blue and C-terminus in red, which was drawn using PDB file 1OPF and PDB Protein Workshop 3.9 (Moreland et al., Citation2005). (ii) Single-crystal neutron diffraction density map of OmpF in tail-deuterated decyl-N,N′-dimethyl amine oxide detergent contrast mapped parallel to the three-fold trimer axis where the porin trimer is represented by the Cα trace (pink) obtained from the X-ray crystal structure. This picture was reproduced with permission from Pebay-Peyroula et al. (Citation1995), which was originally published in Structure (Pebay-Peyroula E, Garavito RM, Rosenbusch JP, Zulauf M, Timmins PA. 1995. Detergent structure in tetragonal crystals of OmpF porin. Structure 3:1051–1059, copyright by Elsevier Inc. 1995. (iii) Structure of the detergent d21-decyl-N,N′-dimethyl amine oxide. The grey area shows the region of deuteration. This Figure is reproduced in colour in the online version of Molecular Membrane Biology.
![Figure 3. Synthesis of d31-LDAO and its use in solubilizing the human voltage-dependent anion channel (VDAC-1) and use of tail-deuterated decyl-N,N′-dimethyl amine oxide in solubilizing Escherichia coli outer membrane protein OmpF. (A) Synthesis of d31-LDAO from d23-dodecanoic acid. The grey areas show the regions of deuteration. (B) (i) [15N-1H]-TROSY spectrum of [U-2H,15N]VDAC-1 in d31-LDAO micelles, (ii) [13C-1H]HMQC spectrum of [U-2H,13C,15N; 1Hδ-IL; 1Hγ-V]VDAC-1 in d31-LDAO micelles highlighting the spectral regions for Ile residues (red box, 11/11 assigned) and Leu plus Val residues (blue box, 8/12 Val and 17/28 Leu assigned), (iii) structure of human VDAC-1 shown as a side view (top) and from above (bottom) with the N-terminus in blue and C-terminus in red, which were drawn using PDB file 2K4T and PDB Protein Workshop 3.9 (Moreland et al., Citation2005). Pictures of the spectra were reproduced with permission from Hiller et al. (Citation2008), which were originally published in Science (Hiller S, Garces RG, Malia TJ, Orekhov VY, Colombini M, Wagner G. 2008. Solution structure of the integral human membrane protein VDAC-1 in detergent micelles. Science 321:1206–1210, copyright by American Association for the Advancement of Science 2008. (C) (i) Crystal structure of the E. coli outer membrane protein OmpF in tetragonal crystal form as an above view with the N-terminus in blue and C-terminus in red, which was drawn using PDB file 1OPF and PDB Protein Workshop 3.9 (Moreland et al., Citation2005). (ii) Single-crystal neutron diffraction density map of OmpF in tail-deuterated decyl-N,N′-dimethyl amine oxide detergent contrast mapped parallel to the three-fold trimer axis where the porin trimer is represented by the Cα trace (pink) obtained from the X-ray crystal structure. This picture was reproduced with permission from Pebay-Peyroula et al. (Citation1995), which was originally published in Structure (Pebay-Peyroula E, Garavito RM, Rosenbusch JP, Zulauf M, Timmins PA. 1995. Detergent structure in tetragonal crystals of OmpF porin. Structure 3:1051–1059, copyright by Elsevier Inc. 1995. (iii) Structure of the detergent d21-decyl-N,N′-dimethyl amine oxide. The grey area shows the region of deuteration. This Figure is reproduced in colour in the online version of Molecular Membrane Biology.](/cms/asset/523c487a-9d51-4978-b6f0-c3e6bb1f75c5/imbc_a_1125536_f0003_c.jpg)
Figure 4. Synthesis of d17- and d24-β-OG and use of d24-β-OG in solubilizing the bacterial outer membrane enzyme PagP. (A) Synthesis of d17- and d24-β-OG from d17-n-octanol and d7-D-glucose. The grey areas show the regions of deuteration. (B) Structure of PagP with the N-terminus in blue and C-terminus in red, which was drawn using PDB file 1MM5 and PDB Protein Workshop 3.9 (Moreland et al., Citation2005). This Figure is reproduced in colour in the online version of Molecular Membrane Biology.
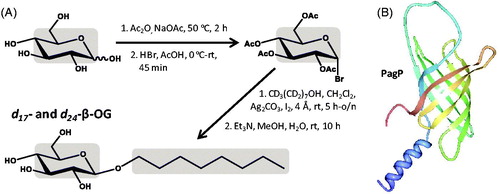
Figure 5. Synthesis of d25- and d39-DDM and use of d39-DDM in solubilizing the E. coli sugar transport protein GalP. (A) Synthesis of d25- and d39-DDM from d25-n-dodecanol and d7-D-glucose, see the work of Hiruma-Shimizu et al. (Citation2014) for the complete synthesis, reaction and experimental details. The grey areas show the regions of deuteration. (B) [15N-1H]TROSY spectrum at 900 MHz of [U-2H, 15N2-Trp]GalP in d39-DDM micelles reproduced from Kalverda et al. (Citation2014) in this journal. This Figure is reproduced in colour in the online version of Molecular Membrane Biology.
![Figure 5. Synthesis of d25- and d39-DDM and use of d39-DDM in solubilizing the E. coli sugar transport protein GalP. (A) Synthesis of d25- and d39-DDM from d25-n-dodecanol and d7-D-glucose, see the work of Hiruma-Shimizu et al. (Citation2014) for the complete synthesis, reaction and experimental details. The grey areas show the regions of deuteration. (B) [15N-1H]TROSY spectrum at 900 MHz of [U-2H, 15N2-Trp]GalP in d39-DDM micelles reproduced from Kalverda et al. (Citation2014) in this journal. This Figure is reproduced in colour in the online version of Molecular Membrane Biology.](/cms/asset/a69eb0f4-41ea-4938-9dd5-02b4116d3677/imbc_a_1125536_f0005_c.jpg)
Figure 6. Synthesis of deuterated fos-cholines and use of d38-DPC in solubilizing the bacterial outer membrane protein OmpA and use of d42-fos-choline-14 in solubilizing the human DAP12-NKG2C complex. (A) Synthesis of deuterated fos-cholines. The grey areas show the regions of deuteration. (B) (i) [15N-1H]TROSY spectrum of OmpA in d38-DPC micelles. This picture was reproduced by permission from Macmillan Publishers Ltd: [Nature Structural Biology] (Arora A, Abildgaard F, Bushweller JH, Tamm LK. 2001. Structure of outer membrane protein A transmembrane domain by NMR spectroscopy. Nat Struct Biol 8:334–338), copyright (2001). (ii) Structure of OmpA with the N-terminus in blue and C-terminus in red, which was drawn using PDB file 1G90 and PDB Protein Workshop 3.9 (Moreland et al., Citation2005). (C) [15N-1H]HSQC spectra of trimer samples segmentally labeled with 15N-2H on the DAP12-only strand (i) or on the DAP12-NKG2C strand (ii) and of the DAP12 homodimer alone (iii) for samples in 250 mM d42-fos-choline-14 with 25 mM d25-SDS and (iv) structure of the DAP12-NKG2C complex with the N-terminus in blue and C-terminus in red, which was drawn using PDB file 2L35 and PDB Protein Workshop 3.9 (Moreland et al., Citation2005). Pictures of the spectra were reproduced by permission from Macmillan Publishers Ltd: [Nature Immunology] (Call ME, Wucherpfennig KW, Chou JJ. 2010. The structural basis for intramembrane assembly of an activating immunoreceptor complex. Nat Immunol 11:1023–1029), copyright (2010). This Figure is reproduced in colour in the online version of Molecular Membrane Biology.
![Figure 6. Synthesis of deuterated fos-cholines and use of d38-DPC in solubilizing the bacterial outer membrane protein OmpA and use of d42-fos-choline-14 in solubilizing the human DAP12-NKG2C complex. (A) Synthesis of deuterated fos-cholines. The grey areas show the regions of deuteration. (B) (i) [15N-1H]TROSY spectrum of OmpA in d38-DPC micelles. This picture was reproduced by permission from Macmillan Publishers Ltd: [Nature Structural Biology] (Arora A, Abildgaard F, Bushweller JH, Tamm LK. 2001. Structure of outer membrane protein A transmembrane domain by NMR spectroscopy. Nat Struct Biol 8:334–338), copyright (2001). (ii) Structure of OmpA with the N-terminus in blue and C-terminus in red, which was drawn using PDB file 1G90 and PDB Protein Workshop 3.9 (Moreland et al., Citation2005). (C) [15N-1H]HSQC spectra of trimer samples segmentally labeled with 15N-2H on the DAP12-only strand (i) or on the DAP12-NKG2C strand (ii) and of the DAP12 homodimer alone (iii) for samples in 250 mM d42-fos-choline-14 with 25 mM d25-SDS and (iv) structure of the DAP12-NKG2C complex with the N-terminus in blue and C-terminus in red, which was drawn using PDB file 2L35 and PDB Protein Workshop 3.9 (Moreland et al., Citation2005). Pictures of the spectra were reproduced by permission from Macmillan Publishers Ltd: [Nature Immunology] (Call ME, Wucherpfennig KW, Chou JJ. 2010. The structural basis for intramembrane assembly of an activating immunoreceptor complex. Nat Immunol 11:1023–1029), copyright (2010). This Figure is reproduced in colour in the online version of Molecular Membrane Biology.](/cms/asset/70253013-ad9a-4721-af38-d53f462c0801/imbc_a_1125536_f0006_c.jpg)
Figure 7. NMR structure of the mitochondrial translocator protein with high affinity ligand determined in d38-DPC micelles. The NMR structure of the mitochondrial translocator protein with high affinity ligand 1-(2-chlorophenyl)-N-methyl-N-(1-methylpropyl)-3-isoquinoline-carboxamide (PK11195) determined in d38-DPC micelles (Jaremko et al., Citation2014) is shown as a side view (i), viewed from the cytoplasm (ii) and viewed from the intermembrane space (iii) with the N-terminus in blue and C-terminus in red, which were drawn using PDB file 2MGY and PDB Protein Workshop 3.9 (Moreland et al., Citation2005). This Figure is reproduced in colour in the online version of Molecular Membrane Biology.
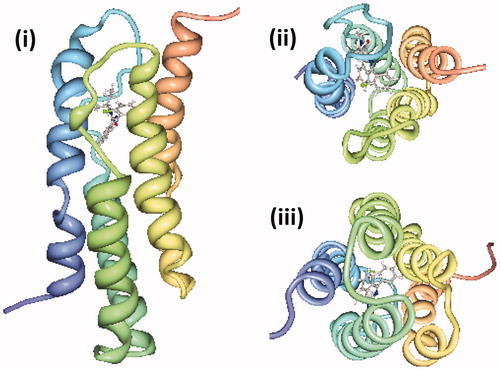
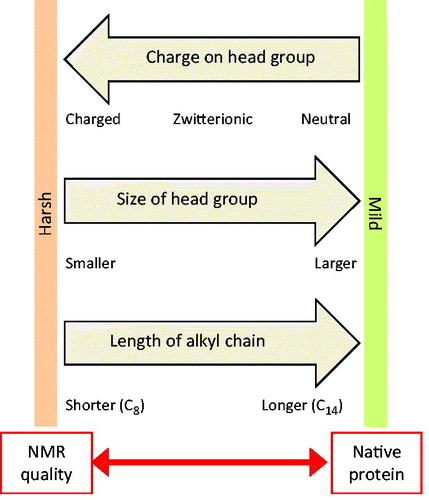
Figure 8. Tensions in selecting a detergent for solution-state NMR structural studies with a membrane protein. This diagram illustrates how the charge on the head group, size of the head group and length of the alkyl group in detergent molecules dictate the properties of detergents and micelle complex formation that provide opposing tensions with regards to achieving good quality NMR spectra and retaining native protein structure and activity. This Figure is reproduced in colour in the online version of Molecular Membrane Biology.