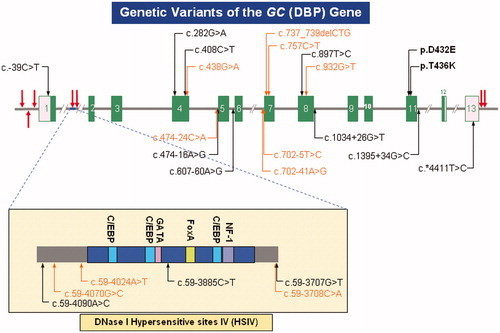
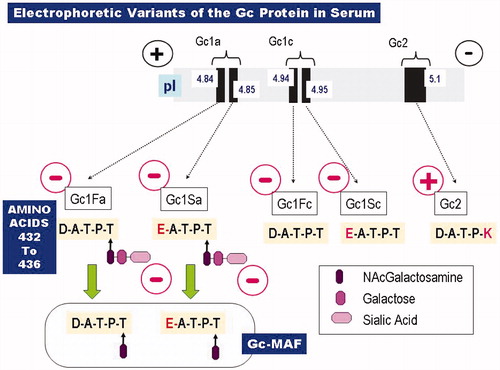
Figures & data
Table 1. Association between common genetic variants of the vitamin D binding protein gene (GC) and various diseases.