Figures & data
Figure 1. Schematic diagram of recombinant pPICZαA harboring HuEpo gene (Epo/PICZαA: 4.1kb). XhoI and XbaI denote to the restriction sites employed for directional cloning of Epo gene into pPICZαA under control of AOX1 promoter, down stream of secretion signal (α-factor) and cleavage sequence (Kex2). AOX1 TT, 6xHis and Stop denote to the transcription termination sequence, His tag and stop codons, respectively.
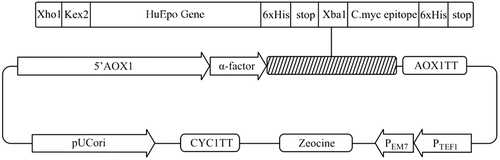
Figure 2. Silver-stained SDS-PAGE analysis of Epo proteins and PEG-PPEpo. Lanes: (1) CHO-derived rHuEpo (R&D Systems); (2) PPEpo; (3) PEGylated PPEpo; (M) Protein size marker SM0431 (Fermentas). The higher apparent MW of CHO-derived rHuEpo (38–39kDa) compared to Pichia-derived rHuEpo (33 kDa) is the result of different glycosylation patterns between two hosts.
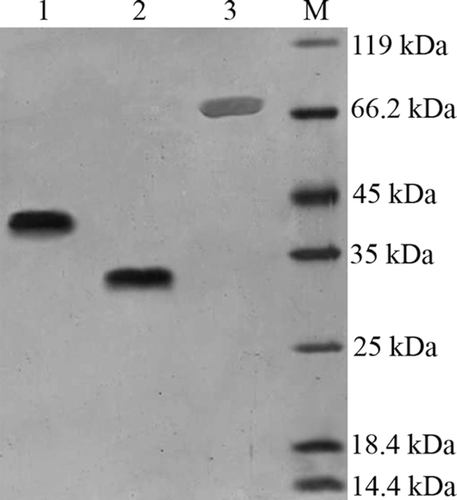
Figure 3. Size-Exclusion HPLC analysis of purified PPEpo and purified PEGylated PPEpo (UV detector at 214 nm). 50 mM sodium phosphate buffer containing 150 mM sodium chloride, pH 7.0, was used as the elution buffer.
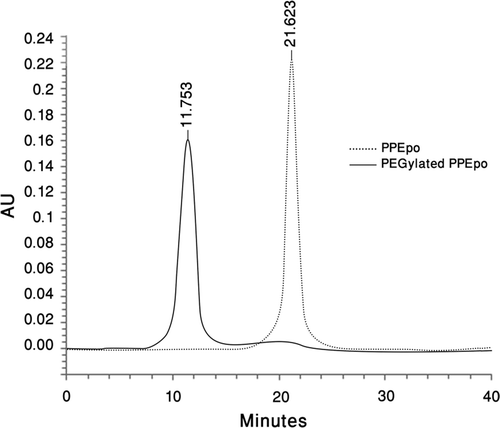
Table 1. The in vitro bioassay results
Figure 4. In vivo activities of rHuEpo (R&D Systems), PPEpo and PEG-PPEpo in the normocythaemic mice. The percentage of reticulocyte count in red blood cells (Ret %) was determined microfluorometrically after subcutaneous administration of each sample at a dose of 10 μg/kg. Data are means ± SD for 4 mice per group.
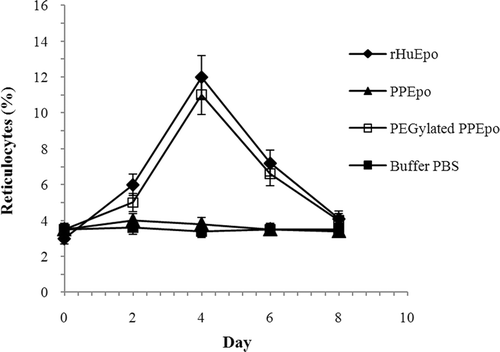
Figure 5. Mean serum concentration vs time profiles of rHuEpo (R&D Systems), PPEpo and PEG-PPEpo following intravenous administration in normal rabbits. Each rabbit received a single 15 μg protein/kg dose. Protein levels were measured using human Epo ELISA kits. Data are means ± SD for 3 rabbits per group.
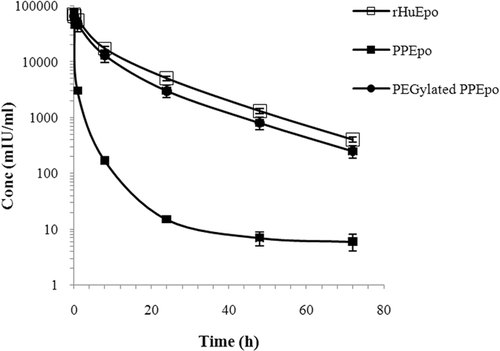
Table 2. A number of pharmacokinetic parameters of rHuEpo, PPEpo and PEGylated PPEpo following intravenous administration in rabbits