Figures & data
Table 1. Bacterial strains and plasmids used in this study
Figure 1. Identification of plasmid pMG36eure and expression of transgene urease in recombinant L. lactis MG1363. A: DNA extracted from recombinant L. lactis MG1363 (lane 1) or recombinant E. coli DH5a (lane 2) were digested with XbaI and HindIII. 5.7 kb and 3.6 kb represent the urease gene and the pMG36e vector. B: The transcript of urease gene was determined by RT-PCR with RNA isolated from recombinant L. lactis MG1363. C, D: Expression of transcriptional protein urease subunits UreE (C), and UreC (D) were analyzed by Western blot using cell extracts from recombinant L. lactis MG1363.
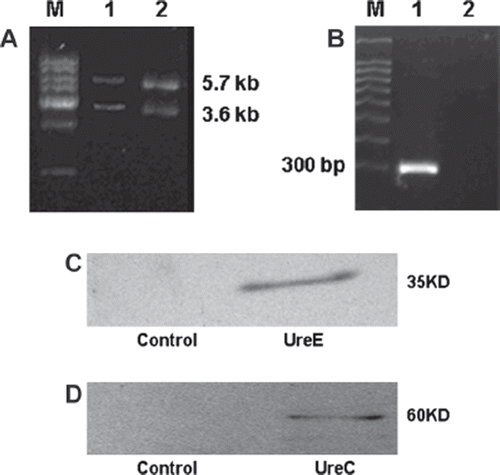
Figure 2. The growth curve of recombinant L. lactis MG1363 and L. lactis MG1363. Strains were grown in GM17 medium at 30°C.
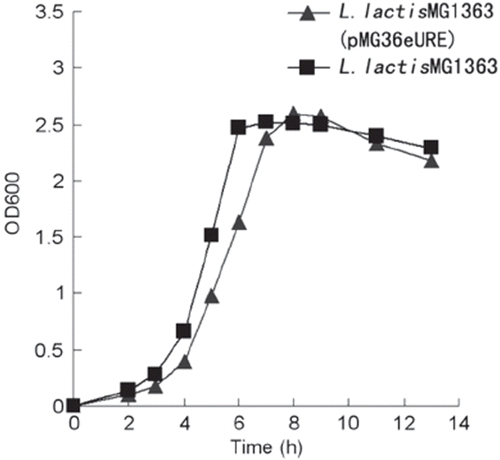
Figure 3. Removal of urea by recombinant L. lactis MG1363 (grown in GM17 medium containing 250μM Ni, with pH maintained at 6.5, 30°C for 24 hours) and E. coli. (grown in LB medium without pH control at 37°C for 12 hours) (n=3).
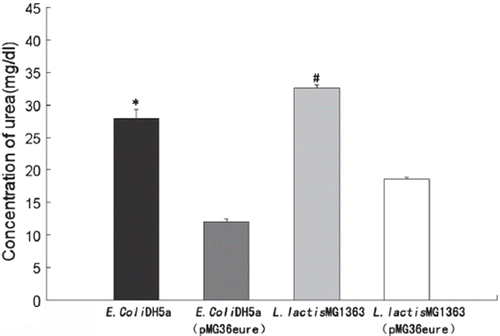
Figure 4. The effect of pH on urea removal. The strains were grown in GM17 medium containing 30mg/dl urea and 250μM NiSO4 at 30°C. The concentration of urea was analyzed after 24h (n=3).
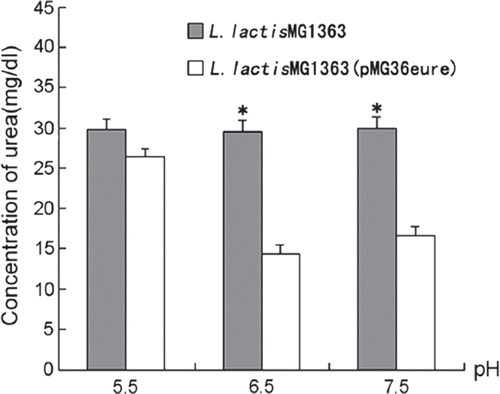
Figure 5. Dependence of urease activity on nickel. Bacteria were grown in GM17 containing 30mg/dl urea supplemented with 0∼800μM NiSO4 at 30°C for 24 hours. The pH was either maintained at 6.5, or without control (n=3).
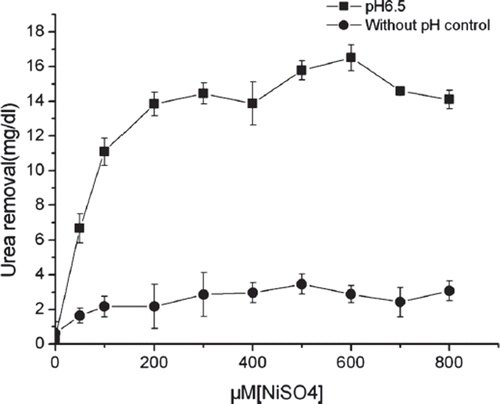
Figure 6. Urea removal in the imitative gastroenteric environment. Cells were first exposed to pH 2.5-4.0 medium for 2 hours, then grown at pH6.8 in a medium containing 0.1 cfu/ml bile acid salt, 30mg/dl urea, and 250μM NiSO4 at 30°C. Control bacteria was exposed to pH4.0 (n=3).
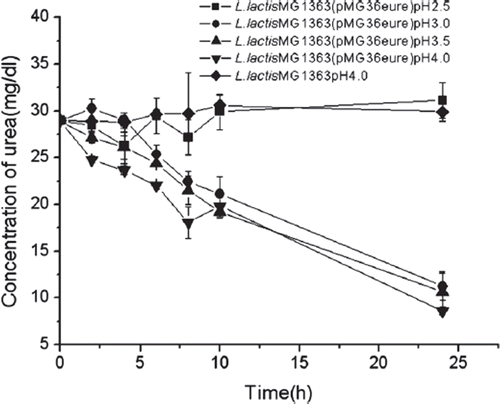
Table 2. Routine blood tests
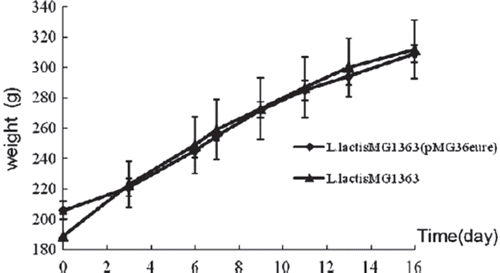
Table 3. Organs weight (% of body weight)
Table 4. Blood biochemical indexes