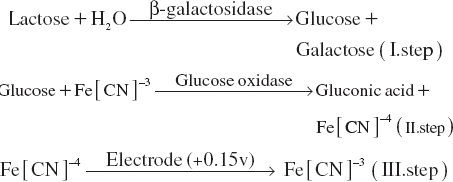Figures & data
Figure 1. Nyquist plot (Zim vs. Zre) for the Faradaic impedance measurements [In all measurements Fe(CN)63−/4−, 0.005 M + 0.1 M KCl, is used as a redox label in the electrolyte solution: (a) A bare glassy carbon electrode. (b) A glucose oxidase-glutaraldehyde-modified electrode. (c) After electropolymerization of anilin. The frequency range is between 0.1 and 100000 Hz with a signal amplitude of 10 mV. Nyquist plots were obtained at a bias potential of 0.17 V vs Ag/AgCl.
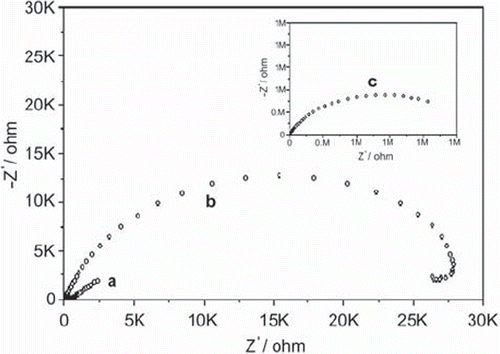
Figure 2. Cyclic voltammograms of (a) bare glassy carbon, (b) glassy carbon/glucose oxidase-glutaraldehyde, (c) glassy carbon/glucose oxidase-glutaraldehyde/polyanilin electrodes in 0.1 M KCl solution containing 5 mM Fe(CN)64−/3−. Scan rate: 50 mV s−1.
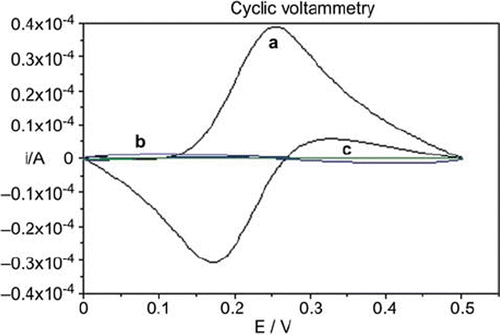
Figure 3. The effect of glucose oxidase activity on the biosensor response [Amounts of glucose activities utilized in biosensors (U): -♦-♦-:45 U, -•-•-:90 U, -▪-▪-:180. Biosensor components: percentages of glutaraldehyde and anilin concentrations were kept constant as 2.5% and 0.4 M, respectively. Electropolymerization potential and polymerization period were 0.6 V and 90 s, respectively. Potential scan conditions: t.puls:40 ms, t.meas:20 ms, U.step:6 mV, scan rate:20 mV s−1. Working buffer was 0.05 M and pH 4.8 citrate solution and contained 1mM ferricyanide as mediator and 0.1 M lactose substrate of β-galactosidase, T = 35°C.]
![Figure 3. The effect of glucose oxidase activity on the biosensor response [Amounts of glucose activities utilized in biosensors (U): -♦-♦-:45 U, -•-•-:90 U, -▪-▪-:180. Biosensor components: percentages of glutaraldehyde and anilin concentrations were kept constant as 2.5% and 0.4 M, respectively. Electropolymerization potential and polymerization period were 0.6 V and 90 s, respectively. Potential scan conditions: t.puls:40 ms, t.meas:20 ms, U.step:6 mV, scan rate:20 mV s−1. Working buffer was 0.05 M and pH 4.8 citrate solution and contained 1mM ferricyanide as mediator and 0.1 M lactose substrate of β-galactosidase, T = 35°C.]](/cms/asset/56d42287-a205-422e-8d9c-8be222188900/ianb19_a_560119_f0003_b.gif)
Table 1. The effects of electropolymerization potential, polymerization peirod, and lactose concentration on the biosensor
Figure 4. The effect of lactose concentration on the biosensor response [Lactose concentrations tested (mM): -▴-▴-:25, -♦-♦-: 50, -•-•-:100, -▪-▪-:150. Biosensor components: Glucose oxidase activity, percentage of glutaraldehyde and anilin concentrations were kept constant as 90 U, 2.5% and 0.4 M, respectively. Electropolymerization potential and polymerization period were 0.6 V and 90 s, respectively. Potential scan conditions: t.puls: 40 ms, t.meas:20 ms, U.step:6 mV, scan rate:20 mV s−1. Working buffer was 0.05 M and pH 4.8 citrate solution and contained 1mM ferricyanide as mediator 0.1 M lactose substrate of β-galactosidase, T = 35°C.]
![Figure 4. The effect of lactose concentration on the biosensor response [Lactose concentrations tested (mM): -▴-▴-:25, -♦-♦-: 50, -•-•-:100, -▪-▪-:150. Biosensor components: Glucose oxidase activity, percentage of glutaraldehyde and anilin concentrations were kept constant as 90 U, 2.5% and 0.4 M, respectively. Electropolymerization potential and polymerization period were 0.6 V and 90 s, respectively. Potential scan conditions: t.puls: 40 ms, t.meas:20 ms, U.step:6 mV, scan rate:20 mV s−1. Working buffer was 0.05 M and pH 4.8 citrate solution and contained 1mM ferricyanide as mediator 0.1 M lactose substrate of β-galactosidase, T = 35°C.]](/cms/asset/36075d2b-6e74-440e-bd17-5de482a2291e/ianb19_a_560119_f0004_b.gif)
Figure 5. β-galactosidase activity graph of the biosensor [Biosensor components: Glucose oxidase activity, percentage of glutaraldehyde and anilin concentrations were kept constant as 90 U, 2.5% and 0.4 M, respectively. Electropolymerization potential and polymerization period were 0.6 V and 90 s, respectively. Potential scan conditions: t.puls:40 ms, t.meas:20 ms, U.step:6 mV, scan rate:20 mV s−1. Working buffer was 0.05 M and pH 4.8 citrate solution and contained 1mM ferricyanide as mediator 0.1 M lactose substrate of β-galactosidase, T = 30°C.]
![Figure 5. β-galactosidase activity graph of the biosensor [Biosensor components: Glucose oxidase activity, percentage of glutaraldehyde and anilin concentrations were kept constant as 90 U, 2.5% and 0.4 M, respectively. Electropolymerization potential and polymerization period were 0.6 V and 90 s, respectively. Potential scan conditions: t.puls:40 ms, t.meas:20 ms, U.step:6 mV, scan rate:20 mV s−1. Working buffer was 0.05 M and pH 4.8 citrate solution and contained 1mM ferricyanide as mediator 0.1 M lactose substrate of β-galactosidase, T = 30°C.]](/cms/asset/e778a04d-ff8d-4eaa-a39d-3295b83005bb/ianb19_a_560119_f0005_b.gif)
Table 2. β-galactosidase activity determination in artificial intestinal juice by the biosensor and by a spectrophotometric reference method.