Figures & data
Figure 1. Diagram of the Whiteside line and transepicondylar axis along with the four point landmarks, namely, the deepest part of the anterior patellar groove (APG), the center of the posterior intercondylar notch (PIN), the lateral epicondylar prominence (LEP) and the median sulcus of the medial epicondyle (MES), conventionally considered to trace the two axes.
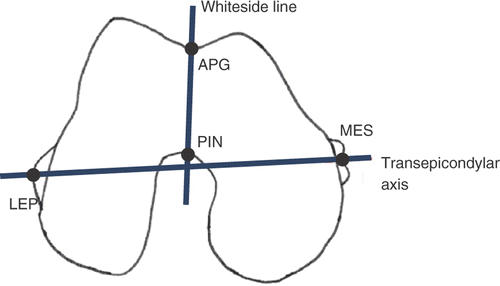
Figure 2. Frontal plane computed through the evolution strategy-based algorithm. The plane orientation is iteratively adjusted through two angles, φ and κ, in the two directions orthogonal to the plane normal .
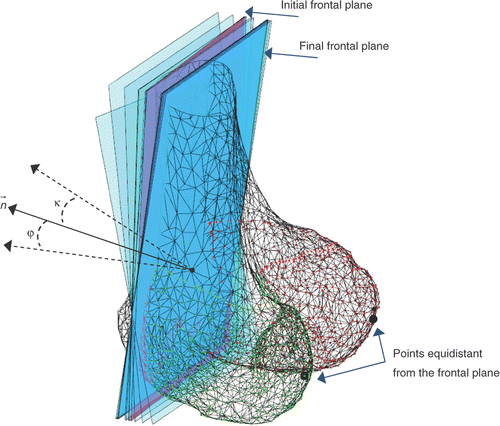
Figure 3. Processing of the intercondylar fossa surface. The frontal plane πf is iteratively shifted in the anterior and posterior direction to obtain the cross-sections of the femur surface. For each cross-section, the algorithm extracts the contour, determines the distal points in the lateral and medial condyles, aligns the 2D profile along the X-axis, separates the 2D profile of the fossa from the overall profile, computes the fifth-order polynomial fitting of the profile, and finally determines the point pc corresponding to the maximum curvature. The anterior margin is automatically detected by using a lower threshold for the maximum curvature of the profile (10% with respect to the maximum curvature measured on the central profiles) as the fossa becomes progressively smoother in the anterior direction. The posterior margin is detected by using a lower threshold (1 mm) on the RMS residual of the polynomial fitting with respect to the profile points. The collected point set {pc} is then used for 3D fitting the WL.
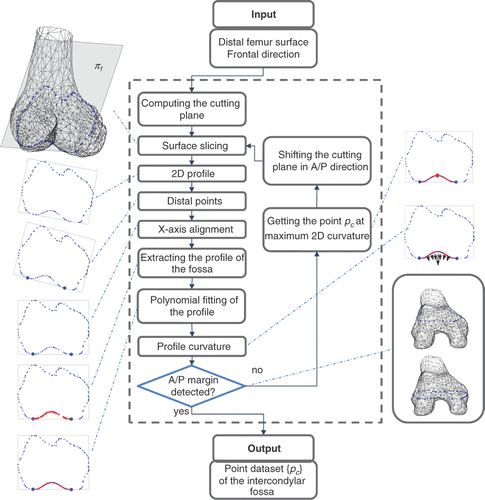
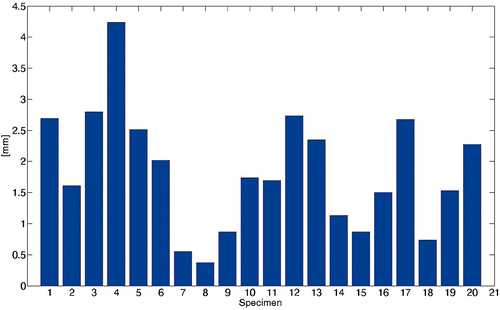
Table I. Fitting error ef (in mm) across the different polynomial orders.
Figure 4. Box-plots of the repeatability results (angular deviation on the axial plane) for the WL. On each box, the central mark is the median, the edges of the box are the 25th and 75th percentiles, and the whiskers extend to the most extreme data points not considered outliers, excluding outliers that are plotted individually as red crosses.
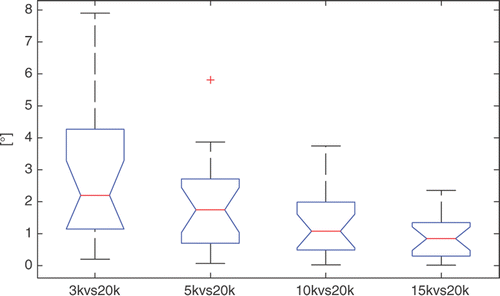
Figure 6. Four representative surfaces of test specimens. The four specimens are (a) #4, (b) #8, (c) #12 and (d) #13 From visual inspection, it is easy to see that the intercondylar fossa of specimen #8 (b) is sharper than those of the other three specimens. As expected, the corresponding 3D fitting error is lower than for the other three specimens.
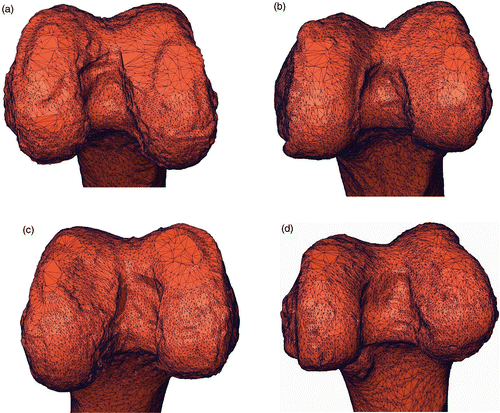
Figure 7. Box-plots of the intra-observer variability {} across the 20 specimens for the four landmarks, namely, the PIN, the APG, the LEP, and the MES, for each of the three expert observers (a, b, c). On each box, the central mark is the median, the edges of the box are the 25th and 75th percentiles, and the whiskers extend to the most extreme data points, excluding outliers that are plotted individually as red crosses.
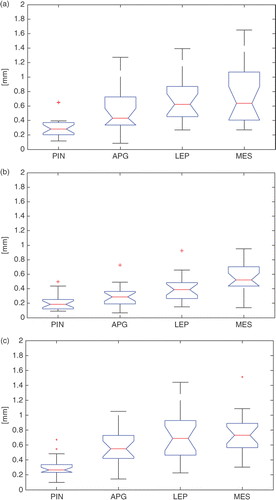
Figure 8. Box-plots of the inter-observer variability for the four landmarks (PIN, APG, LEP, MES) and the corresponding WL and TEA. On each box, the central mark is the median, the edges of the box are the 25th and 75th percentiles, and the whiskers extend to the most extreme data points, excluding outliers that are plotted individually as red crosses.
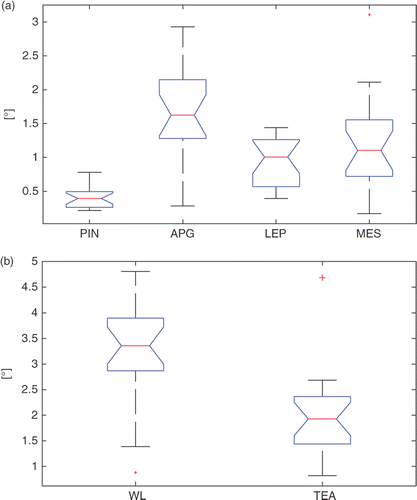
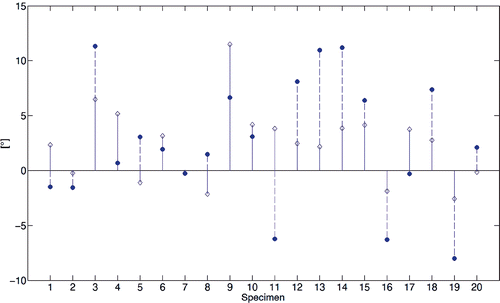
Figure 9. Stem diagram of the angular skew between the two WL axes computed by virtual palpation and with the automated method.
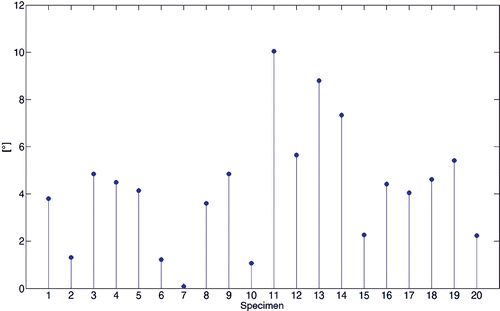
Figure 10. Stem diagram of the perpendicularity between the WL axes and the TEA. Diamond and dot marks refer to the WL computed by the automated method and the virtual palpation, respectively. Values greater than 0° indicate an external rotation of the WL relative to the TEA; smaller values indicate a relative internal rotation.