Figures & data
Figure 1. Parameters related to the XFEM enrichment Heaviside function corresponding to a 3D crack surface.
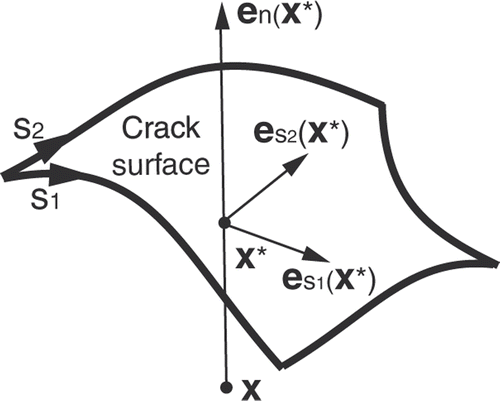
Figure 2. Segmentation of intraoperative images. (1a) Original second iMR image, acquired before retraction. (2a) Original third iMR image, acquired after retraction. (1b) Whole-brain region segmented out manually from (1a) with 3D Slicer. (2b) Whole-brain region segmented out manually from (2a) with 3D Slicer. (1c) Smoothing of the segmented whole-brain region from (1b). (2c) Smoothing of the segmented whole-brain region from (2b). (1c) and (2c) are performed to minimize the dependence of the active surface algorithm on segmentation roughness and surface mesh resolution.
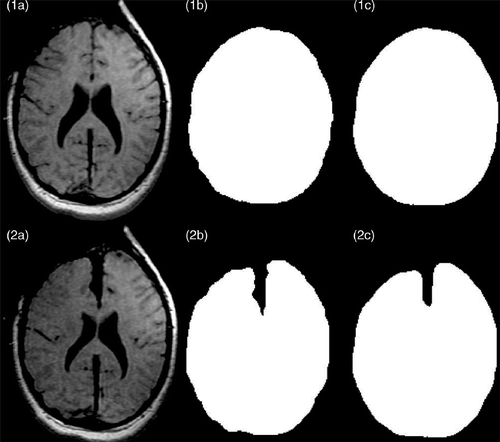
Figure 3. Representation of the planar cut to be included in the biomechanical model. (a) Surface mesh of the biomechanical model showing the location of the plane where the retractor was inserted. This plane corresponds to the plane separating the two hemispheres. (b) Whole-brain region segmented out from the second iMR image, acquired before retraction, in the selected plane shown in (a). (c) Whole-brain region segmented out from the the third iMR image, acquired after retraction. (d) Cut defined as the difference between (b) and (c), where (c) has been processed beforehand to remove small, isolated islands. (e) Surface mesh representing the intersections of the cut with the volume mesh of the whole-brain region brain.
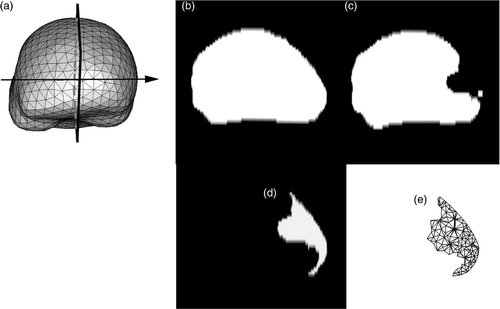
Figure 4. Results of the active surface algorithm for the two hemispheres of the brain. (a) Initial active surface for right hemisphere with color levels corresponding to the magnitude of the displacement field. (b) Initial active surface for left hemisphere with same color coding. (In accordance with radiological convention, the right side of the iMR images corresponds to the left side of the brain, and vice versa.
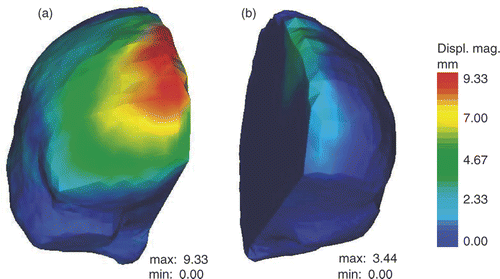
Figure 5. Evaluation of the displacement field of the cut for 3D retraction modeling. (a) Whole-brain region segmented out from the third iMR image, with segment (in red) defining the angle of the displacement field of the right lip. (b) Surface mesh built from the segmented third iMR image, cut mesh () (in red), and displacements of cut intersections (in green). (Displacements go from right to left in the figure.) (c) Cut mesh () with color levels corresponding to the magnitude of the displacement field of the right lip.
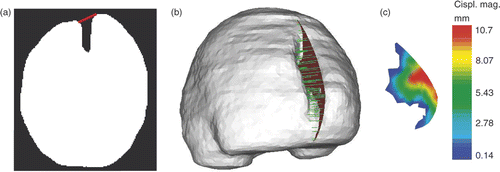
Figure 6. Comparison of unsmoothed (row 1) and jointly smoothed (row 2) displacement fields of right hemisphere and right lip of the cut. (1a) Surface meshes of right hemisphere and right lip of cut, and their surface displacement fields in green. (1b) Same as (1a), except that the surface meshes are left transparent. (1c) Zoom of (1b) around the external surface of the brain. (2a), (2b) and (2c) are, respectively, the same as (1a), (1b) and (1c), except that the displacement fields are jointly smoothed. (In each sub-figure, the direction of the displacement vectors is defined such that the vectors start at the surface mesh nodes.)
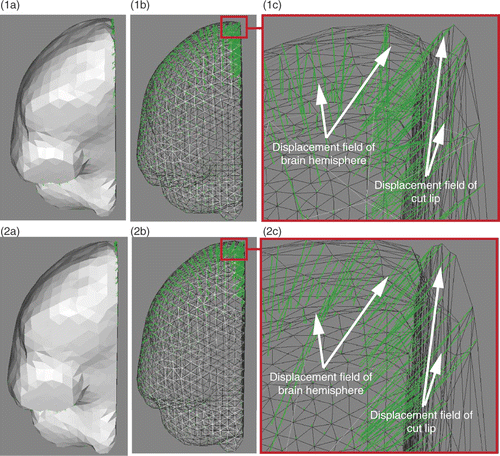
Figure 7. Deformation of the biomechanical model based on XFEM for retraction modeling. (a) External surface mesh of the biomechanical model with color levels corresponding to the magnitude of the displacement field. (b) Slice of the biomechanical model with same color coding. (c) Final mesh resulting from the modeling of the retraction using XFEM. The tetrahedra that were added to display the opening of the cut lips are there only for visualization purposes. The edges of FEs cut by the discontinuity have actually been made discontinuous and their nodes moved apart.
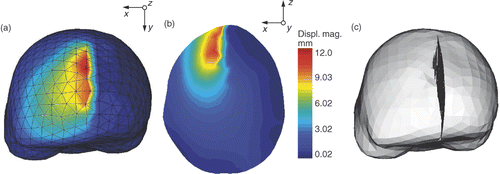
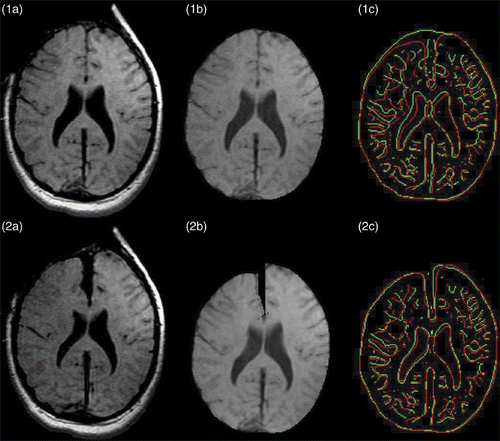
Figure 8. Results of retraction modeling. (1a) Original second iMR image. (2a) Third iMR image, rigidly registered to (1a). (1b) Whole-brain region extracted from (1a). (2b) Deformation of (1b) using the volume displacement field of the biomechanical model, computed via XFEM. (1c) Juxtaposition of edges of whole-brain region of second iMR image (1a) (in green) and edges of whole-brain region of third iMR image (2a) (in red). (2c) Juxtaposition of edges of whole-brain region of second iMR image deformed for modeling retraction (2b) (in green) and edges of whole-brain region of third iMR image (2a) (in red). (The red edges in (1c) and (2c) are identical.)