Figures & data
Figure 1. 3D visualization of the vertebra used in the experiments. The dots are examples of points selected on the surface of the vertebra that will be used to generate PCpatient. Blue dots are points visible from the current viewing direction, and black dots are those occluded by the tip of the spinous process.
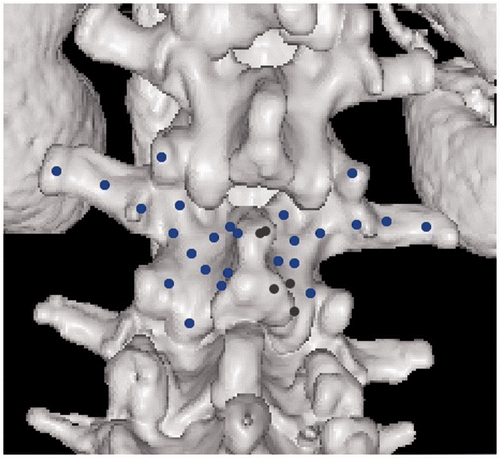
Figure 2. Illustration of the sampling line, indicated by a red line on two orthogonal sections (left and center) and a 3D model (right).
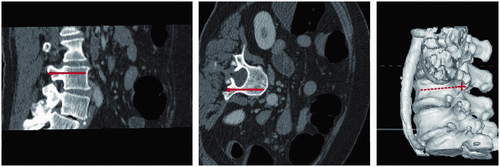
Table I. Number of points falling in each zone for the six PCpatient for studying the influence of point distribution on the accuracy of surface matching.
Figure 3. SRE with different searching coefficients (N) of global registration. The dots represent the mean SRE, and the bars indicate the standard error.
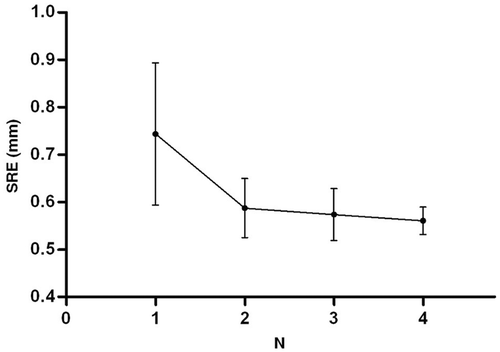
Figure 4. TRE for the sampling points with different searching coefficients (N) of global registration. The dots represent the mean TRE, and the bars indicate the upper bound of the 95% confidence intervals.
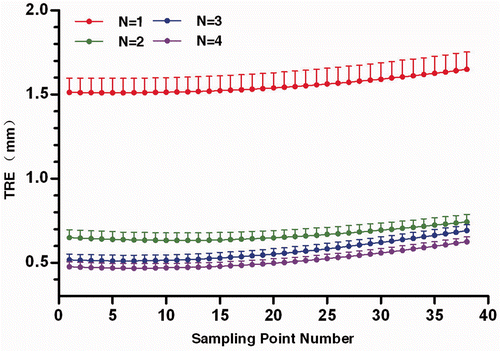
Figure 5. TRE for the sampling points with six different PCpatient as listed in . The dots represent the mean TRE, and the bars indicate the upper bound of the 95% confidence intervals.
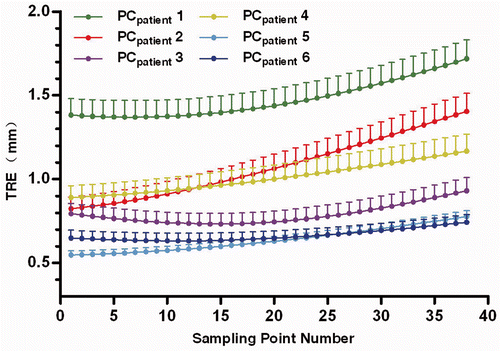
Table II. Statistical significance of the difference between the TRE for the sampling points 1, 13, 25 and 38 of PCpatient 1, 2, 3, 4, 5 and that of PCpatient 6.
Figure 6. SRE with different number of points in PCpatient. The dots represent the mean SRE, and the bars indicate the standard error.
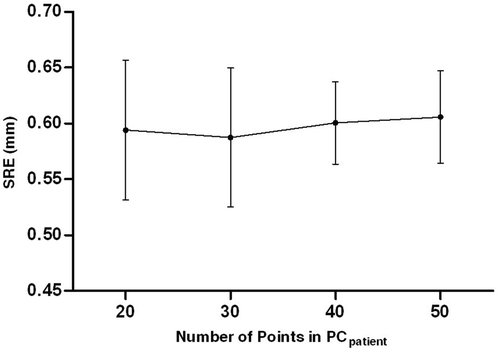
Figure 7. TRE for the sampling points with different number of points (C) in PCpatient. The dots represent the mean TRE, and the bars indicate the upper bound of the 95% confidence intervals.
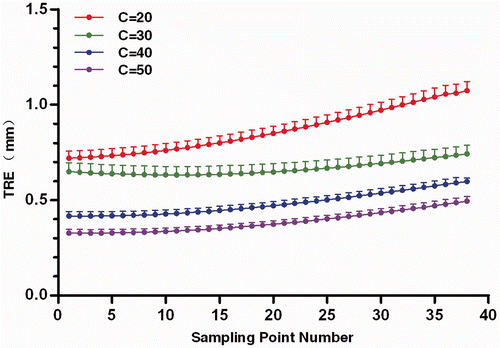
Table III. Total number of points and their distribution in different zones for studying the influence of point number on the accuracy of surface matching.