Figures & data
Table 1. Recipe for qindan-capsule formulation.
Table 2. Quality evaluation of the qindan-capsule compound.
Table 3. Primers for RT-PCR.
Figure 1. Identification of AFs: (A) AFs cells (100×); (B) negative expression of SMA by immunocytochemistry (100×); and (C) positive expression of vimentin by immunocytochemistry (100×).
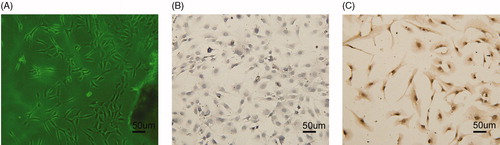
Figure 2. The effects of drug-containing serum on AF migration. AFs were treated with TGF-β1 for 24 h. Cell migration was measured with the transwell assay. The values shown represent the mean ± SEM, n = 3. &, p < 0.01 compared to NC; #, p < 0.05 compared to PC; *, p > 0.05 compared to losartan; Student’s t-test.
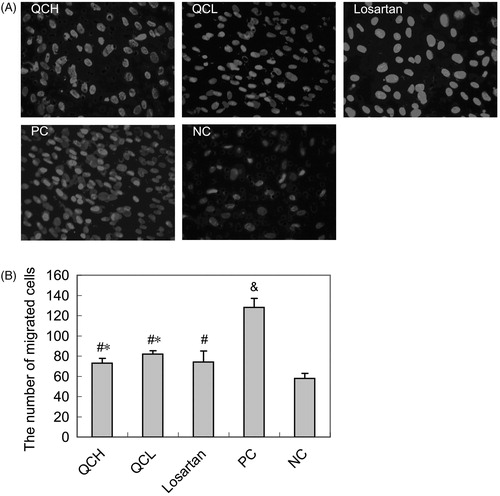
Figure 3. The effects of drug-containing serum on AF proliferation. AFs were treated with TGF-β1 for 24 h. Cell growth was measured with the MTT assay. The values shown represent the mean ± SEM, n = 3. &, p < 0.01 compared to NC; #, p < 0.05 compared to PC; ##, p < 0.01 compared to PC; *, p > 0.05 compared to losartan; **, p < 0.05 compared to losartan; Student’s t-test.
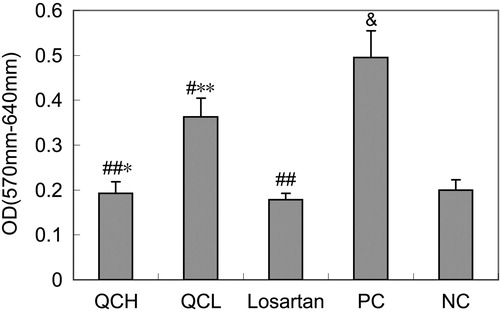
Figure 4. The effects of drug-containing serum on the AF cell cycle. AFs were treated with TGF-β1 for 24 h. Cell cycle progression was evaluated by flow cytometry. The values shown represent the mean ± SEM, n = 3. &, p < 0.01 compared to NC; $, p < 0.05 compared to NC; &&, p > 0.05 compared to NC; ##, p < 0.05 compared to PC; #, p > 0.05 compared to PC; *, p > 0.05 compared to losartan; Student’s t-test.
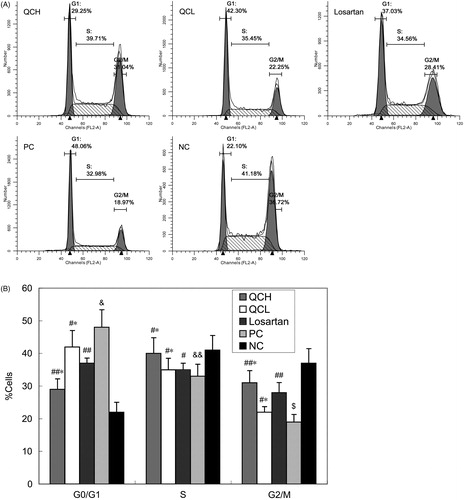
Figure 5. The effects of drug-containing serum on AF apoptosis. AFs were treated with TGF-β1 for 24 h. Cell apoptosis was measured by flow cytometry. (A) Representative data shows apoptosis cells. (B) Columns, mean of data obtained from three independent experiments. Bar mean SEM. &, p < 0.01 compared to NC; ##, p < 0.05 compared to PC; #, p < 0.01 compared to PC; *, p > 0.05 compared to losartan; Student’s t-test.
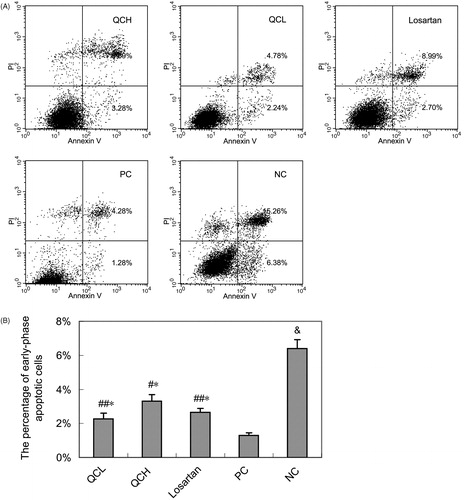
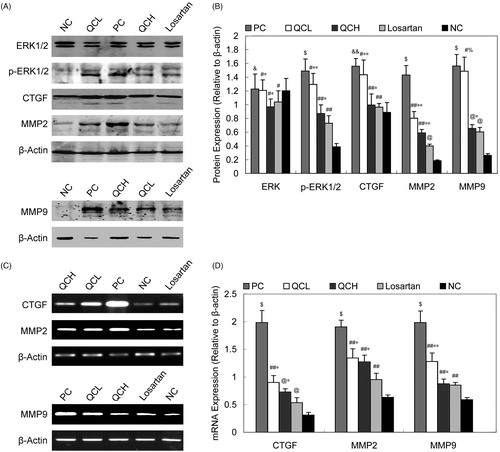
Figure 6. ERK1/2, CTGF, MMP2 and MMP9 expression. (A) and (B): protein levels of ERK1/2, p-ERK1/2, CTGF, MMP2 and MMP9. The values shown represent the mean ± SEM, n = 3. &&, p < 0.05 compared to NC; $, p < 0.01 compared to NC; &, p > 0.05 compared to NC; #, p > 0.05 compared to PC; ##, p < 0.05 compared to PC; @, p < 0.01 compared to PC; *, p > 0.05 compared to losartan; **, p < 0.05 compared to losartan; %, p < 0.01 compared to losartan; Student’s t-test. (C) and (D): CTGF, MMP2 and MMP9 mRNA expression. The values shown represent the mean ± SEM, n = 3. $, p < 0.05 compared to NC; $, p < 0.01 compared to NC; &&, p > 0.05 compared to NC; ##, p < 0.05 compared to PC; #, p > 0.05 compared to PC; *, p > 0.05 compared to losartan; **, p < 0.05 compared to losartan; Student’s t-test.