Figures & data
Figure 1. Total phenol (g/L), flavonol (mg/L), and flavan-3-ol (mg/L) contents of latex and CCS. Note: CCS was produced by treatment of CL phosphate buffer. n.e. = not evaluated.
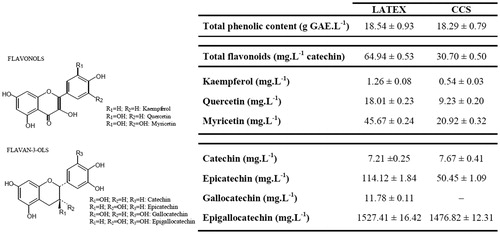
Figure 2. Effects of CCS and the PC doxorubicin at 31.25–500 µg/mL on cell viability of EAC and MCF-7 cells. All data are expressed as mean ± SD, n = 3. Significant differences between treatment groups and the NC group are denoted by ** and ***, indicating p < 0.01 and p < 0.001, respectively.
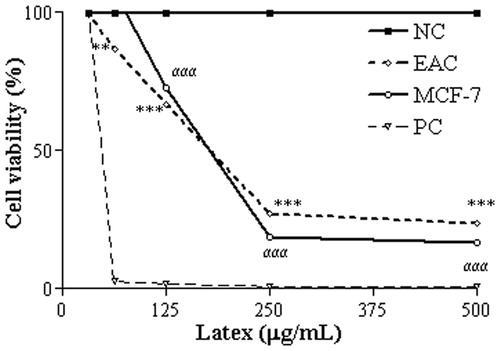
Figure 3. Damage profile to plasmid DNA after treatment with CCS at 0.78–25 µg/mL Statistically significant differences (p < 0.001) in the abundance of FI, FII and FIII plasmid forms between the CSS treatments and the NC are denoted by α***, β*** and γ***, respectively, (n = 3). NC, NC treated with saline; PC, PC plasmid linearized with EcoRI. Tests were performed in triplicate and results are expressed as Mean ± SD.
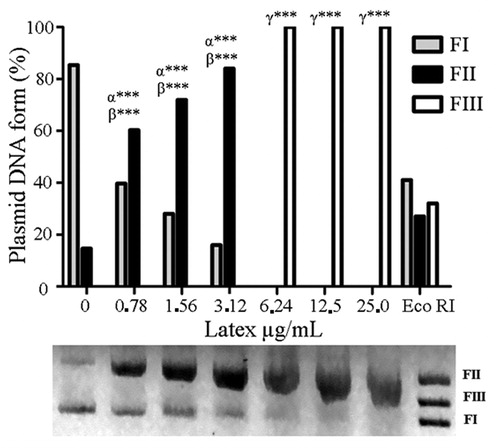
Figure 4. Cell death indicated by EB/AO differential staining of EAC cells after 48 h treatment with CCS at 169 µg/mL. Data are expressed as mean ± SD, n = 3. Statistically significant differences (p < 0.001) between treatments and NC and PC (62.5 µg/mL doxorubicin) are denoted by α*** and β***, respectively, (n = 3).
Figure 5. Inhibition of tumor growth by CCS treatments at 0.78, 1.56 and 3.12 mg/mL, and in the PC (62.5 µg/mL doxorubicin). Data are expressed as mean ± SD, n = 6. Statistically significant differences (p < 0.001) between treatments and NC and PC are denoted by α*** and β***, respectively.
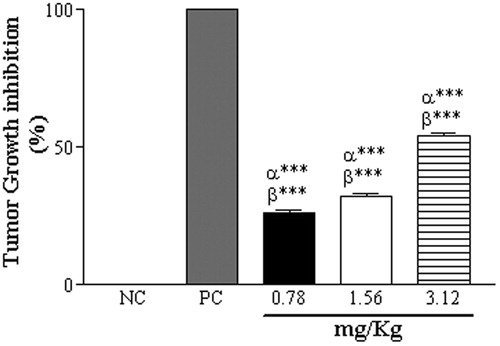
Table 1. Effect of CCS treatment at doses of 0.78 (C1), 1.56 (C2) and 3.12 mg/kg/day (C3), and PC (62.5 µg/mL doxorubicin) on morphologic parameters of EAC-bearing mice.