Figures & data
Table 1. Phytocomponents identified in the ethanol leaf extract of MA by GC-MS analysis.
Table 2. IC50 values of MA compared with that of standard ascorbic acid.
Figure 2. Hydroxyl radical scavenging effect of MA with different concentrations in comparison with standard ascorbic acid. Values are given as mean ± SD of six replicates in each group. Bar values are sharing a common superscript (a,b,c) differ significantly at p < 0.05 DMRT.
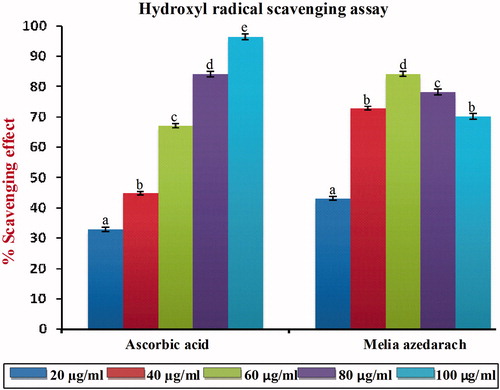
Figure 3. Superoxide radical scavenging effect of MA with different concentrations in comparison with standard ascorbic acid. Values are given as mean ± SD of six replicates in each group. Bar values are sharing a common superscript (a,b,c) differ significantly at p < 0.05 DMRT.
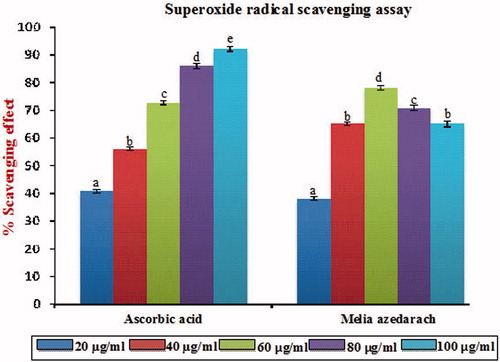
Figure 4. Nitric oxide radical scavenging effect of MA with different concentrations in comparison with standard ascorbic acid. Values are given as mean ± SD of six replicates in each group. Bar values are sharing a common superscript (a,b,c) differ significantly at p < 0.05 DMRT.
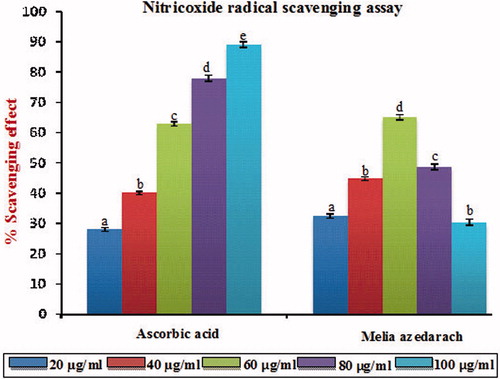
Figure 5. DPPH radical scavenging effect of MA with different concentrations in comparison with standard ascorbic acid. Values are given as mean ± SD of six replicates in each group. Bar values are sharing a common superscript (a,b,c) differ significantly at p < 0.05 DMRT.
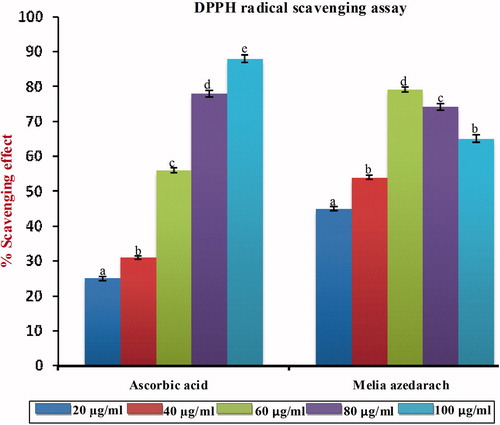
Figure 6. Free radical scavenging activity of MA with different concentrations in comparison with standard ascorbic acid as measured in reducing power assay. Values are given as mean ± SD of six replicates in each group. Bar values are sharing a common superscript (a,b,c) differ significantly at p < 0.05 DMRT.
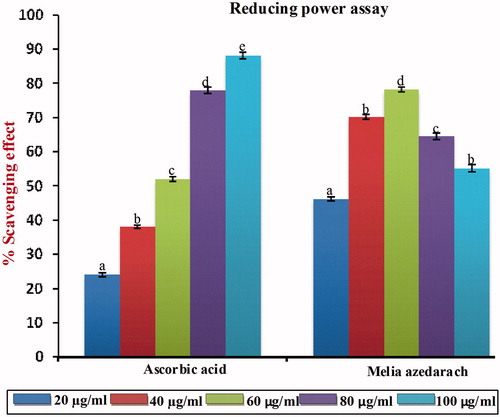
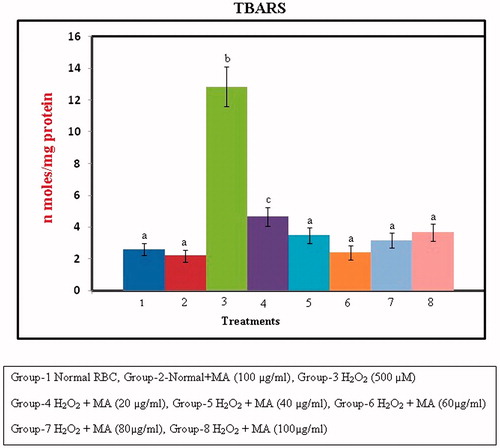
Figure 7. Protection against H2O2-induced lipid peroxidation in RBC cellular membrane by different concentrations of MA. Values are given as mean ± SD of six replicates in each group. Bar values are sharing a common superscript (a,b,c) differ significantly at p < 0.05 DMRT.
Figures 8. Changes in the levels of DNA damage (% DNA in tail, tail length, tail moment and Olive tail moment) in cultured lymphocytes on pretreatment with ethanol extract of MA. Values are given as mean ± SD of six replicates in each group. Bar values are sharing a common superscript (a,b,c) differ significantly at p < 0.05 DMRT.
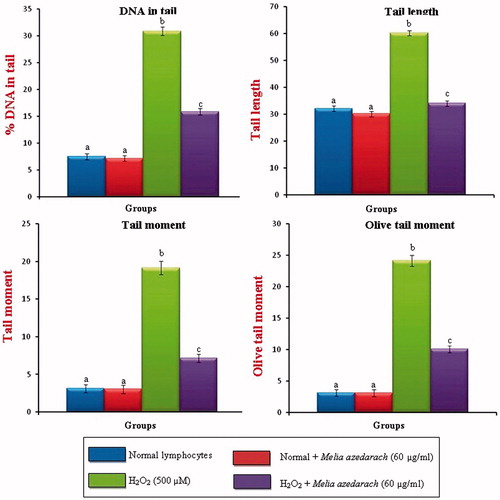
Figure 9. Changes in the levels of DNA damage in normal, H2O2 and Melia azedarach pretreated lymphocytes.
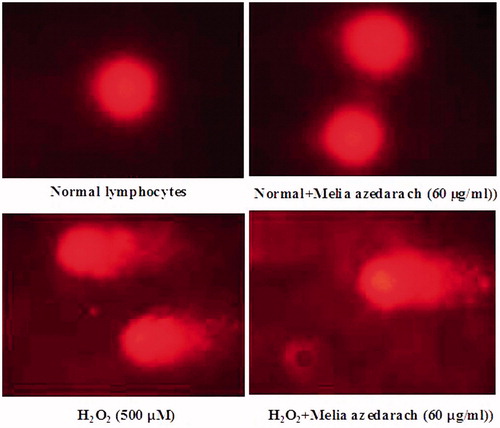
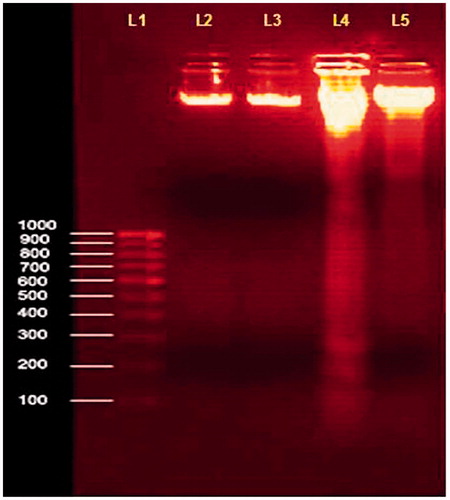
Figure 10. Changes in the DNA fragmentation assay in cultured lymphocytes on pretreatment with ethanol extract of MA. Values are given as mean ± SD of six replicates in each group. Lane (L1)-1000bP ladder DNA; L2-Normal lymphocytes; L3-Normal + MA (60 µg/mL); L4-H2O2 (500 µM); L5-H2O2 + MA (60 µg/mL).