Figures & data
Figure 1. Phragmites australis partial aro A gene amplification. The PCR amplification of partial aro A gene is shown in lanes 2 and 3. A 1 Kb MW marker is shown in lane 1.
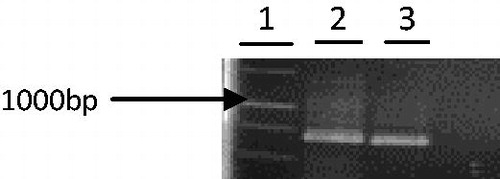
Figure 2. Analysis of the pTZ carrying the aro A gene. The PCR-amplified insert of the plasmid is shown in lanes 3–5 and plasmid without insert (negative control) is shown in lane 2. A 1 kb MW marker is shown in lane 1. Plasmid of lane 3 was selected to sequence.
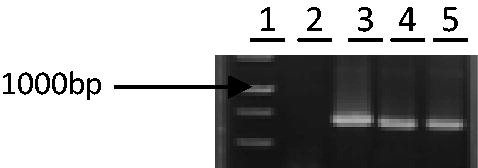
Figure 3. The partial sequence of Phragmites australis with intron regions. The underlined regions are primers (forward (TKZF) and reverse (TKZR)). The regions in gray box are introns (86 bp and 289 bp).

Figure 4. The partial sequence of aro A of Phragmites australis without intron regions. The underline regions are primers (forward (TKZF) and reverse (TKZR)).

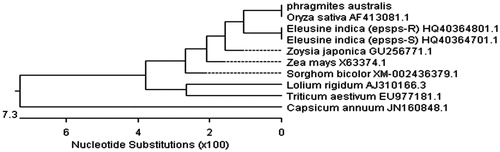
Figure 5. The phylogenic relationship of Phragmites australis with other species. A phylogenic tree (unrooted) based on the genetic distance of amino acid sequences was constructed by the Clustal method with using the DNAstar software.