Figures & data
Figure 1. Effect of JEE on LPS-stimulated NO/PGE2 production and iNOS/COX-2 expression in RAW 264.7. (A) The levels of nitrite and protein expression of iNOS were determined by Griess reagents and Western blotting, respectively. (B) PGE2 production and COX-2 protein expression were determined by EIA and Western blotting, respectively. The data presented are means ± SEM of three independent experiments. *p < 0.05, **p < 0.01, and ***p < 0.001 were used to indicate significance compared with the LPS-stimulated value.
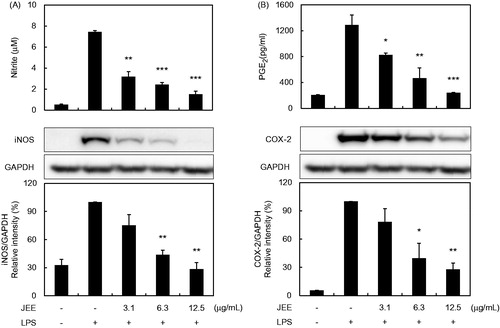
Figure 2. Effect of JEE on LPS-stimulated production and mRNA expression of IL-1β and IL-6 in RAW 264.7 cells. (A) The production of IL-1β and IL-6 was determined by ELISA. (B) Total mRNA was prepared and transcription levels in the cells were detected by a real-time (RT)-PCR analysis. The data presented are means ± SEM of three independent experiments. *p < 0.05, **p < 0.01, and ***p < 0.001 were used to indicate significance compared with the LPS with the LPS-stimulated value.
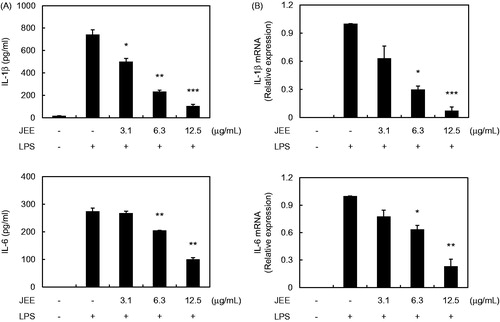
Figure 3. Effect of JEE on LPS-stimulated phosphorylation of IκBα and NF-κB in RAW 264.7 cells. (A) Cell lysates were analyzed by Western blotting analysis using specific antibodies against p-IκBα, IκBα, and NF-κB. (B) To confirm the effect of JEE on NF-κB activation, the binding activity of NF-κB to DNA was determined in nuclear extract by Trans-AM p65 assay. *p < 0.05, **p < 0.01, and ***p < 0.001 were used to indicate significance compared with the LPS with the LPS-stimulated value.
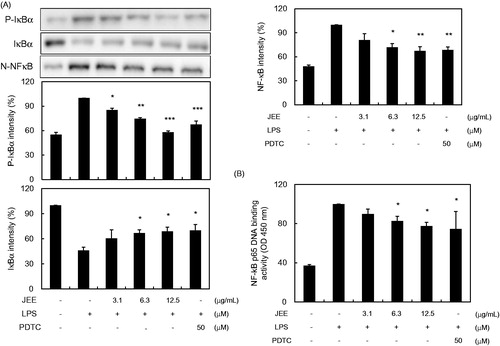
Figure 4. Effect of JEE on LPS-stimulated activation of MAP kinases in RAW 264.7 Cells. Cell lysates were prepared and analyzed by Western blotting using antibodies against p-ERK, p-JNK p-38, ERK, JNK, and p38. The data presented are means ± SEM of three independent experiments. *p < 0.05, **p < 0.01, and ***p < 0.001 were used to indicate significance compared with the LPS-stimulated value.
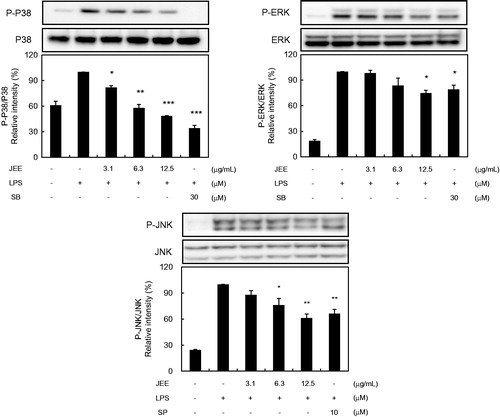
Figure 5. Effect of JEE on TPA-induced ear edema of mice. JEE and indomethacin were applied topically immediately after TPA treatment, and ear thickness measured at before and 1.5, 3, 4.5, and 6 h intervals. Each value represents as means ± SEM. *p < 0.05, **p < 0.01, and ***p < 0.001 were used to indicate significance compared with control.
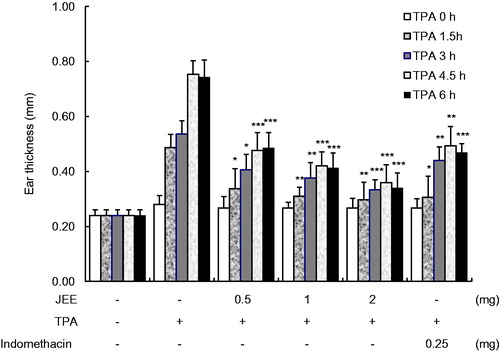
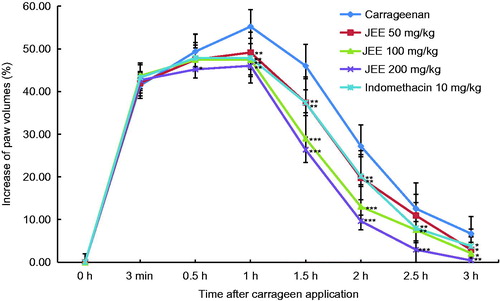
Figure 6. Effect of JEE on carrageenan-induced paw edema of mice. JEE and indomethacin were administered orally 30 min before carrageenan injection into the left hind paw, and paw volume measured at 3 min, 0.5, 1, 1.5, 2, 2.5, and 3 h intervals after carrageenan injection. Each value represents as means ± SEM. *p < 0.05, **p < 0.01, and ***p < 0.001 were used to indicate significance compared with control.