Figures & data
Figure 1. Analysis of growth inhibitory effect of RC, MM, and RD on LNCaP cells. (A) Chemical structures of RC, MM, and RD. (B) Summary of IC50 values of RC, MM, and RD in LNCaP cells. (C) The effect of 24 h treatment of RC, MM, and RD on growing LNCaP cells was determined by MTT assay. Control cells were treated with equal volumes of DMSO supplemented in medium. ***p < 0.001 compared with the Mib+ control group. (D) After exposure of LNCaP cells to RC, MM, or RD for 24 h, cells were harvested, and stained with propidium iodide, and cell cycle was analyzed by flow cytometry. (E) Percentage of cell populations in cell cycle. (F) Percentage of apoptotic cells. ***p < 0.001 compared with the Mib+ control group. (G) Western blotting analysis of p53, p21Cip1, E2F-1, Bcl2, Bax, and PARP in the cells after RC, MM, or RD treatment for 24 h.
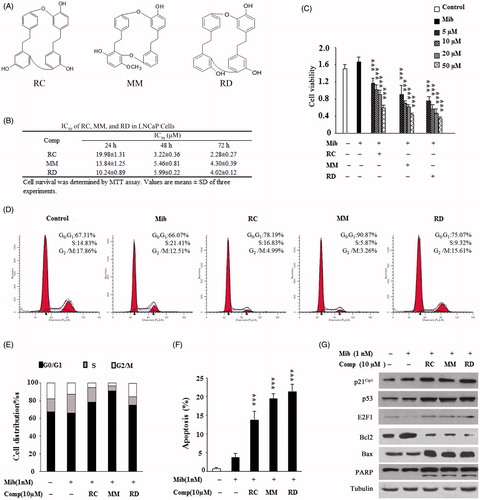
Figure 2. Analysis of AR expression in LNCaP after treated with RC, MM, or RD. (A) Quantitative PCR analysis of AR mRNA levels. ***p < 0.001 compared with the Mib+ control group. (B) AR promoter activity analysis by relative luciferase activity assays after treatment with the chemicals for 1 h and 24 h respectively. *p < 0.05, **p < 0.005, and ***p < 0.001 compared with the control group. (C) Western blotting analysis of AR.
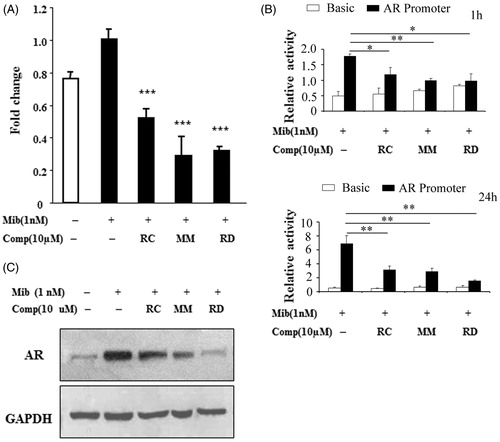
Figure 3. The impact of RC, MM, and RD on AR transcriptional activity. Endogenous (A) and exogenous (B) AR transcriptional activities were analyzed by detecting the activity of the promoter of PSA. ***p < 0.001 compared with the control group. Endogenous (C) and exogenous (D) AR transcriptional activities were analyzed by detecting the activity of ARE. *p < 0.05 and ***p < 0.001 compared with the control group. (E) ELISA analysis of PSA in LNCaP cell media after treated with RC, MM, or RD. ***p < 0.001 compared with the control group. (F) Representative immunofluorescence staining of AR (Red) in LNCaP cells with/without compounds treatment. Nuclei were counterstained with DAPI (blue). (G) Cytoplasmic and nucleoplasmic levels of AR were detected by western blot after treated with RC, MM, or RD. (H) Associations of AR and SRC-1 were determined by coimmunoprecipitation using anti-AR.
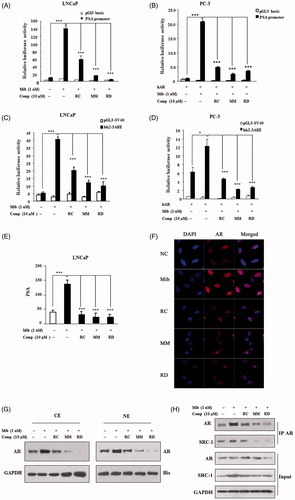
Figure 4. RC, MM, and RD repress proteasome activity. (A) The compounds were used to directly act on purified 20S proteasome, and proteasome activities were detected. Columns, mean (n = 3); bars, SD; ***p < 0.001 compared with the control group (DMSO treatment). (B) LNCaP cells were treated with the compounds for 24 h, and cell lysates were performed to detect proteasome activities. Columns, mean (n = 3); bars, SD; **p < 0.005 and ***p < 0.001 compared with the control group. (C) Western blotting analysis of ubiquitinated protein in LNCaP cells after treated with RC, MM, or RD. (D) Polyubiquitinated-AR and polyubiquitinated-p21Cip1 were detected by coimmunoprecipitation using anti-HA after transfection of a HA-ubiquitin expression plasmid in LNCaP cells.
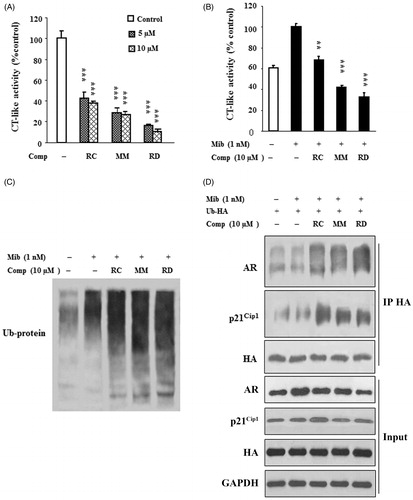
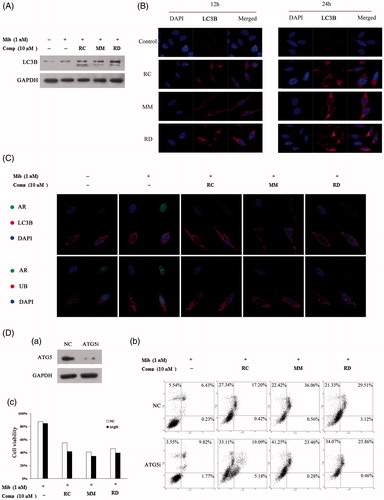
Figure 5. Induction of autophagy by RC, MM, or RD in LNCaP cells. (A) Western blot analysis of LC3 in compound-treated LNCaP cells. (B) Representative images of the punctuated LC3 in LNCaP cells treated with RC, MM, or RD. (C) Representative immunofluorescence staining of AR (green) and LC3 or Ub (red) in compound-treated LNCaP cells. Nuclei were counterstained with DAPI (blue). (D) (a) Western blotting was performed to confirm ATG5 knockdown; (b) percentage of apoptotic cells was analyzed by flow cytometry after ATG5 knockdown; (c) cell viability assays after depletion of autophagy using Atg5-targeting siRNA in LNCaP cells prior to treatment with the chemicals.