Figures & data
Figure 1. The structure of ecotin (PDB ID: 1ECZCitation17). In (a) a ribbon diagram of the antiparallel homodimer is shown with protease interaction sites 1 and 2 indicated. In (b) a single monomer is shown with the residues mutated in this paper illustrated in space-filling format. The diagram was made using PyMol (www.pymol.com).
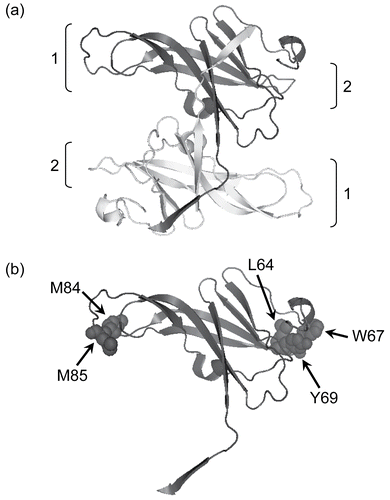
Figure 2. (a) The expression and purification of recombinant ecotin. The recombinant protein is indicated by an arrow and runs close to the 18 kDa marker, as expected. (i) Extraction of a periplasmic extract. Uninduced and induced refer to cell extracts immediately before and 3 h after induction of the cells with IPTG. Osmotic shock refers to a sample of the bacterial suspension following treatment with osmotic shock buffer, and the last lane is the periplasmic extract. (ii) Purification by ion-exchange chromatography. The periplasmic extract was loaded onto a ResourceQ column (see “Materials and methods”). Ecotin washed through the column under the conditions of the experiment whereas most of the impurities remained bound. Periplasm refers to a sample of the periplasmic proteins prior to application to the ion-exchange column. Flow through refers to samples of fractions from the column. The fractions marked with asterisks were used without further purification. (b) Recombinant ecotin inhibits trypsin (▪ and solid line), elastase (▴ and dotted line), and chymotrypsin (▾ and dashed line). To enable the data to be shown on a single graph, the rates were normalized such that the rate in the absence of ecotin was equal to 1.0. Each point represents the mean of three separate determinations, the error bars the standard error of these means, and the lines the non-linear curve fit obtained as described in “Materials and methods.”
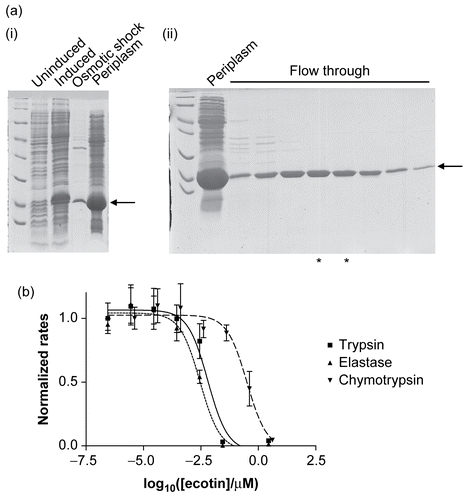
Figure 3. The effects of alanine mutations on the interaction between ecotin and (a) trypsin, (b) chymotrypsin, and (c) elastase. In each case, the negative logarithm of the IC50 is plotted as a bar which represents the mean of three separate determinations; the error bars represent the standard deviations of these means.
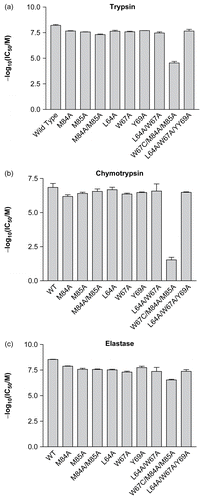
Table 1. Experimentally determined IC50 values (nM) for wild-type and mutant ecotin with trypsin, chymotrypsin, and elastase.
