Figures & data
Figure 1. (a) The reaction catalyzed by N-acetylgalactosamine kinase. (b) The active site of GALK2. Only the substrates and key residues are shown. The loop comprising Thr-184 to Gly-186 accommodates the N-acetyl group of N-acetylgalactosamine (GalNAc). In galactokinase, the threonine and glycine residues in this loop are substituted for bulkier methionine and cysteine side chains respectively. Asp-190 is located with the carboxyl group in between the C1-OH of the sugar and the γ-phosphate of the nucleotide. This residue is likely to be important in the catalytic mechanism of the enzyme and its ionization state is probably influenced by Arg-43. The figure was drawn using PyMol (www.pymol.org) and the PDB file 2A2DCitation8. (c) Structures of N-acetylgalactosamine kinase inhibitors used in this study.
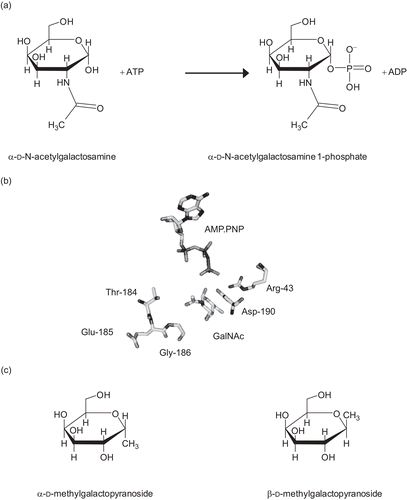
Figure 2. Purification of recombinant human GALK2. Samples taken during the purification procedure (see methods) were analyzed by 10% SDS-PAGE. The sizes of molecular mass markers (M) are shown on the left. U, extract from cells prior to induction; I, extracts from induced cells prior to harvesting by centrifugation; S, protein solution extracted from cells by sonication and clarified by centrifugation; F, material which passed through the nickel–agarose column; E, protein eluted from this column in the presence of 250 mM imidazole.
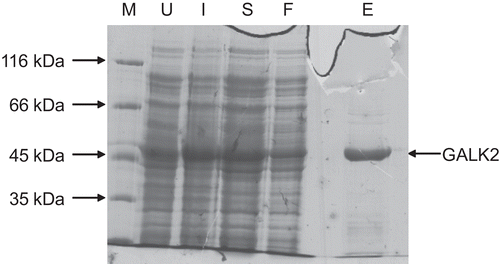
Figure 3. Recombinant human GALK2 (50 nM) is an active N-acetylgalactosamine kinase. In (a), the concentration of ATP was maintained at a constant level of 0.5 mM and N-acetylgalactosamine (GalNAc) was varied. The resulting data were fit (i) to the Michaelis–Menten equation (giving values of Km,GalNAc = 40 ± 14 μM and kcat = 1.0 ± 0.1 s−1) and (ii) to a modified version of this equation which accounts for substrate inhibition (giving values of Kis = 13 ± 8 mM, Km = 73 ± 27 mM, and kcat = 1.2 ± 0.1 s−1). In (b) the GalNAc concentration was maintained at 5 mM and the concentration of ATP was varied (giving values of Km,ATP = 14 ± 3 μM and kcat = 1.0 ± 0.1 s−1). Each point represents the mean of two or three separate determinations and the error bars the standard deviations of these means.
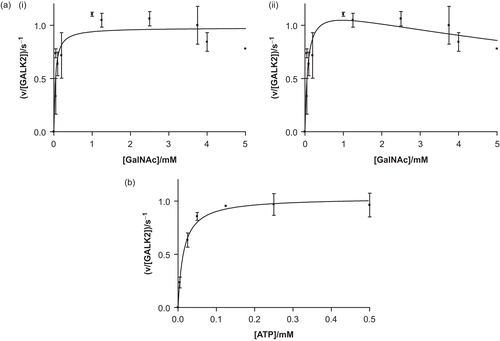
Figure 4. The inhibition of GALK2 (50 nM) by α-methylgalactopyranoside. Inhibition was studied over a range of α-methylgalactopyranoside concentrations and three different ATP concentrations: 8 μM (inverted triangles, ▾), 15 μM (squares, ▪), and 60 μM (upright triangles, ▴). (a) In a Dixon plot the lines do not converge, suggesting that the inhibition is not competitive. (b) The converging lines on a plot of [ATP]/rate against [inhibitor] suggest that the mode of inhibition is uncompetitive with respect to ATP. In all cases the N-acetylgalactosamine concentration was 1.0 mM.
![Figure 4. The inhibition of GALK2 (50 nM) by α-methylgalactopyranoside. Inhibition was studied over a range of α-methylgalactopyranoside concentrations and three different ATP concentrations: 8 μM (inverted triangles, ▾), 15 μM (squares, ▪), and 60 μM (upright triangles, ▴). (a) In a Dixon plot the lines do not converge, suggesting that the inhibition is not competitive. (b) The converging lines on a plot of [ATP]/rate against [inhibitor] suggest that the mode of inhibition is uncompetitive with respect to ATP. In all cases the N-acetylgalactosamine concentration was 1.0 mM.](/cms/asset/4421913e-ef71-49d6-a963-7cc1546b1749/ienz_a_418122_f0004_b.gif)
Figure 5. The effects of pH on the turnover number, kcat. The GALK2 concentration was 230 nM in these experiments. The line represents a fit to the equation kcat = kcat,lim(1 + [H+]/Ka1 + Ka2/[H+]).
![Figure 5. The effects of pH on the turnover number, kcat. The GALK2 concentration was 230 nM in these experiments. The line represents a fit to the equation kcat = kcat,lim(1 + [H+]/Ka1 + Ka2/[H+]).](/cms/asset/40afba64-ba34-42a4-81c2-328ad8ef9dbe/ienz_a_418122_f0005_b.gif)
