Figures & data
Figure 2. Effects of compounds on cellular NOX activity. (A) Representative kinetic traces of cytochrome c reduction by PMA-stimulated human neutrophils alone (black circles) or in the presence of either SOD (white circles) or 100 µM DPI (gray circles). (B) Prior to stimulation with phorbol ester, human neutrophils were incubated in the presence of perhexiline (IC50 = 3.0 ± 0.2 µM, Hill slope = −3.1 ± 0.9), Shionogi 1 (IC50 = 56 ± 3 nM, Hill slope = −1.4 ± 0.1), Shionogi 2 (IC50 = 99 ± 6 nM, Hill slope = −1.8 ± 0.2), VAS2870 (IC50 = 77 ± 14 nM, Hill slope = −1.0 ± 0.2), DPI (IC50 = 39 ± 5 nM, Hill slope = −1.1 ± 0.1), or suramin (inactive). NOX activity was determined using detection of SOD-inhibitable cytochrome c reduction as described in “Methods”.
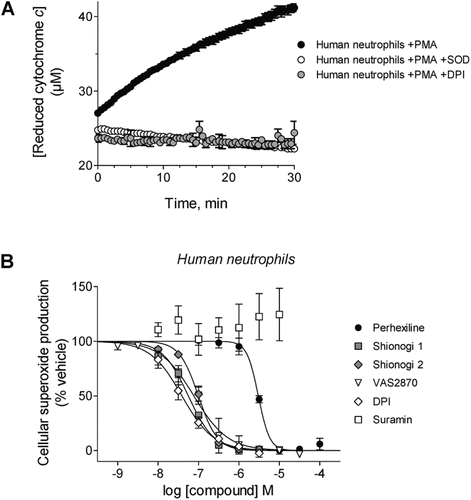
Figure 3. Protein components of the semi-recombinant NOX2 assay. (A) Representative anti-gp91phox immunoblot analysis of neutrophil membranes used in the semi-recombinant assay. A characteristic broad band centered at ~91 kD was observed when the membranes were denatured at 70°C for 5 min (left), consistent with literature reports for glycosylated gp91phox (see, for example, ref 35. Denaturation at 100°C for 10 min elicited a single band at ~60 kD (right), consistent with deglycosylation of the protein (theroretical MW = 65.3 kD) as a result of sample preparation. (B) Representative gel images of purified, recombinant, His-tagged cytosolic proteins p47phox (left), p67phox (middle) and Rac2 (right). For each protein, the Coomassie-stained gel (2 µg of protein) and the corresponding immunoblot (40 ng of protein) are shown.
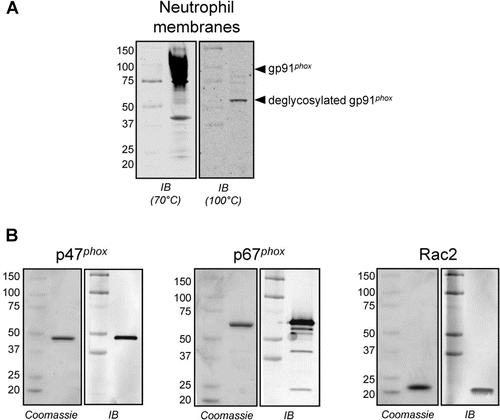
Figure 4. Effects of compounds on purified semi-recombinant NOX2 activity. (A) Representative kinetic traces of cytochrome c reduction by NADPH-stimulated semi-recombinant NOX2 alone (black circles) or in the presence of either SOD (white circles) or 2 µM DPI (gray circles). (B) Prior to reaction initiation with NADPH, neutrophil membranes and recombinant cytosolic components were added to perhexiline (IC50 = 13 ± 2 µM, Hill slope = −0.75 ± 0.08), Shionogi 1 (inactive), Shionogi 2 (inactive), suramin (IC50 = 1.0 ± 0.1 µM, Hill slope = −1.0 ± 0.1), VAS2870 (inactive), or DPI (IC50 = 90 ± 10 nM, Hill slope = −0.84 ± 0.07). NOX2 activity was determined using SOD-inhibitable cytochrome c reduction as described in “Methods”.
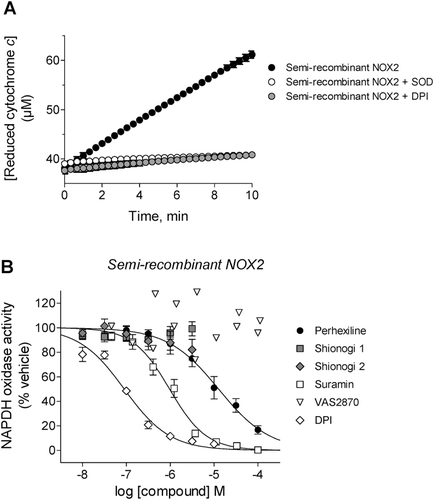
Figure 5. Testing for inhibitory effects of compounds by NOX2-independent mechanisms. (A) Inhibition of xanthine oxidase activity by perhexiline (inactive) and luteolin (a positive control) was measured using SOD-inhibitable cytochrome c reduction as described in “Methods”. (B) Inhibition of PKCβII activity by cellular-active inhibitors Shionogi 1 (IC50 = 4.6 ± 0.8 nM, Hill slope = −1.0 ± 0.1), Shionogi 2 (IC50 = 9.4 ± 1.0 nM, Hill slope = −1.0 ± 0.1), perhexiline (inactive) and VAS2870 (inactive) as well as sangivamycin (a positive control, IC50 = 6.8 ± 2.0 µM, Hill slope = −0.7 ± 0.1), was determined using a fluorescence polarization IMAP™ kit as described in “Methods”.
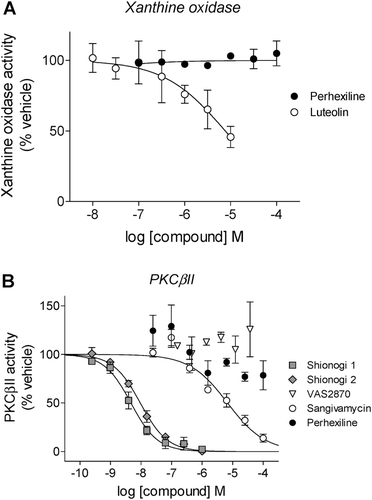
Figure 6. Inhibition of p47phox translocation in PMA-stimulated neutrophils: representative immunoblots (top) and observed intensities of the p47phox bands (normalized to the intensity of the corresponding band for the integral membrane component of NOX2, gp91phox), relative to the PMA-treated samples from corresponding gels (bottom). Neutrophils were treated with vehicle, PMA (1 µM), or PMA following pretreatment with apocynin, VAS2870, perhexiline, suramin, Shionogi 1, or Shionogi 2, then membrane fractions were isolated and analyzed by Western analysis as described in “Methods”. With the exception of apocynin (100 µM), all compounds were tested at 10 µM. The order of sample lanes (top) correspond to the order of intensity bars (bottom).
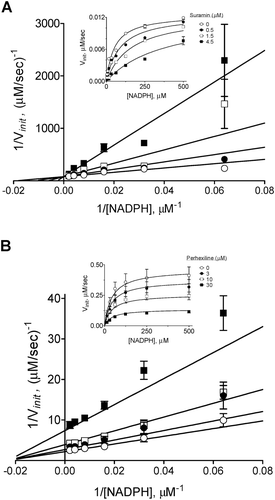
Figure 7. Mode of NOX2 inhibition by suramin (A) and perhexiline (B). Inhibition modalities were determined by global nonlinear fitting and model comparison as described in “Methods”. Lineweaver-Burk plots (foreground) were generated by reciprocal transformation of the best nonlinear fits (inset). Solid lines represent the best-fit global trends. (A) Suramin exhibits a competitive pattern of inhibition with respect to NADPH (best-fit parameters: Ki = 0.7 ± 0.1 µM, Km = 52 ± 8 µM, Vmax = 0.013 ± 0.001 µM/s). (B) Perhexiline exhibits a noncompetitive pattern (best-fit parameters: Ki = 13 ± 2 µM, Km = 44 ± 8 µM, Vmax = 0.46 ± 0.03 µM/s). Data shown are representative; differences in Vmax values between the two data sets reflect the typical variability observed with neutrophil membrane preparations.