Figures & data
Figure 1. Ribbon representation of the crystal structure of C. albicans Secreted aspartic protease 2, Sap2, (PDB code: 1EAG). Residues in contact with A70450 are shown in cyan.
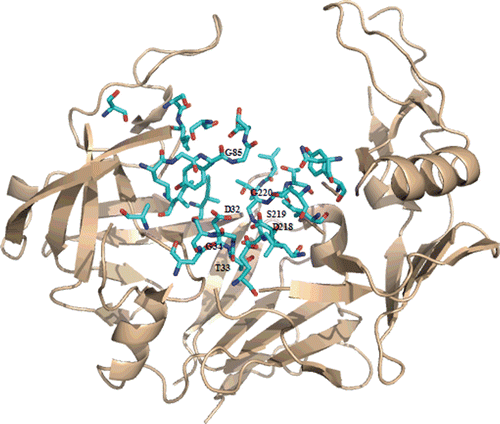
Table 1. Inhibition potency of FDA HIV-1 aspartic protease inhibitors towards Sap2 and HIV-1 protease.
Figure 2. Ribbon representation of the crystal structure of HIV-1 protease (PDB code: 3OXC). Residues in contact with saquinavir are shown in cyan.
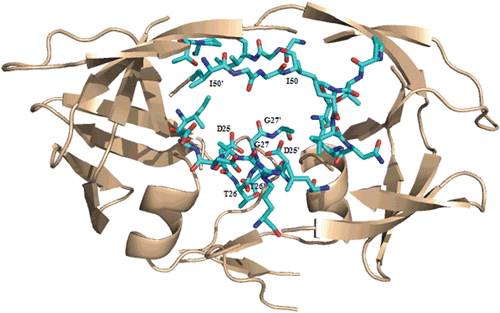
Figure 3. Sequence comparison between Sap2 (1EAG) and HIV-1 aspartic protease (3OXC) in the active site region performed using EMBOSS matcher pairwise sequence alignment: Sap2 N-terminal domain, top; C-terminal domain, bottom.
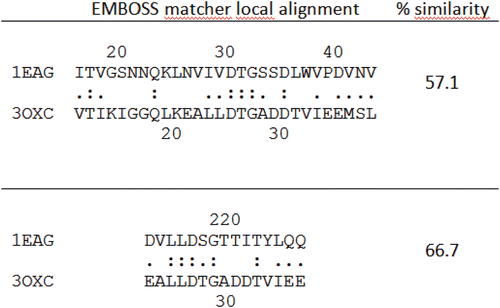
Figure 4. Top: superimposition of HIV-1 protease (PDB: 1OXC in light pink) and C.albicans Sap2 (PDB: 1EAG in green) obtained with SPDBV. Down: binding pockets superimposition.
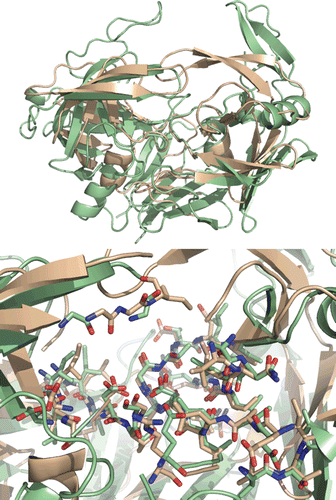
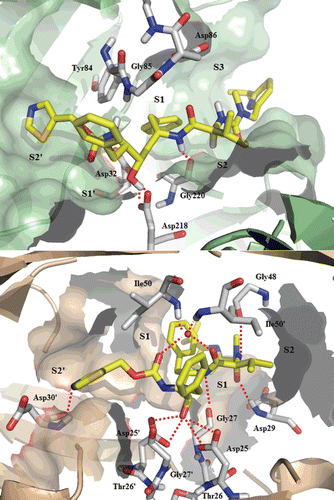
Figure 5. Top: Best-scoring docked conformations resulting from the docking of saquinavir into Sap2 binding site. Down: saquinavir in complex with HIV-1 protease (PDB code: 3OXC).
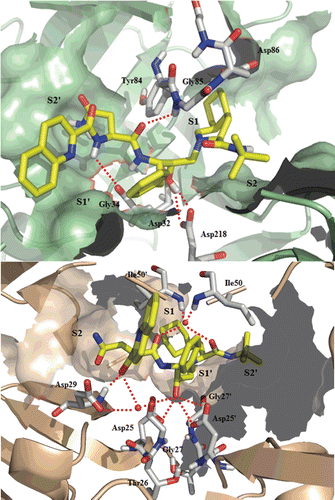
Figure 6. Top: Best-scoring docked conformations resulting from the docking of ritonavir into Sap2 binding site. Down: ritonavir in complex with HIV-1 protease (PDB code: 1HXW).