Figures & data
Figure 1. Schematic of the characterized enzyme constructs, with the protease domain (black), the helicase domain (dark grey), the co-factor NS4A (medium grey) and N-terminal His-tags (light grey).
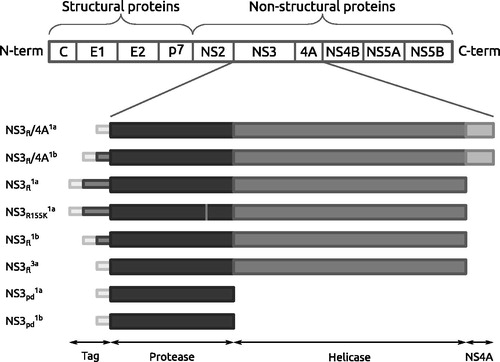
Figure 3. Illustration of the relationship between the enzymatic activity of and the concentration of OGP.
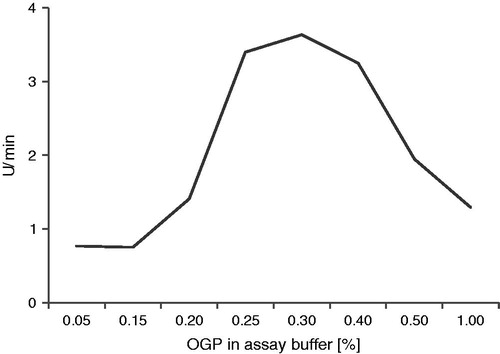
Table 1. Reference and optimized buffer compositions for analysis of proteolytic activity of NS3 variants.
Figure 4. Dependence of enzyme activity on detergent, glycerol and NaCl concentration for the studied enzyme variants. (a) in 0.4% OGP; (b)
in 0.4% OGP; (c)
in 0.4% OGP; (d) NS3fl/4A1a without NS4Apep in 0.4% Triton; (f) NS3 fl/4A1a with NS4Apep in 0.4% Triton; (e) NS4 fl/4A1b without NS4Apep in 0.4% Triton; (g) NS3 fl/4A1b with NS4Apep in 0.4% Triton; (h)
in 0.7% OGP; (i)
in 0.3% OGP; (j)
in 0.4% OGP.
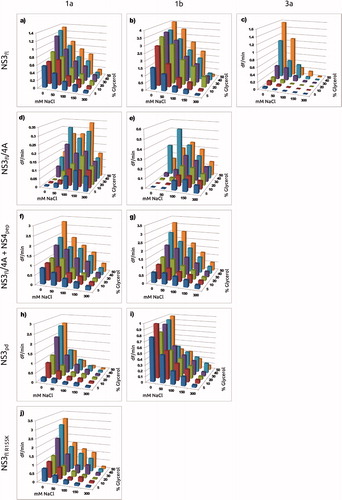
Table 2. Catalytic parameters of NS3 protease variants in reference and optimized buffers.
Table 3. Inhibition constants (Ki) for NS3 protease variants in reference and optimized buffers.
Table 4. Vitality values for the four inhibitors in this study under optimized conditions, where is used as reference.