Figures & data
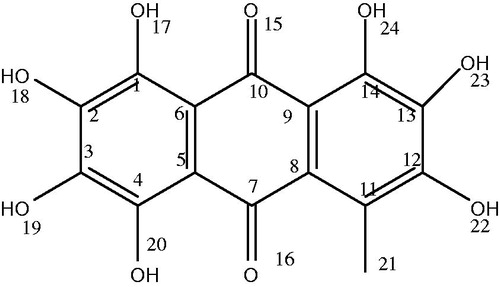
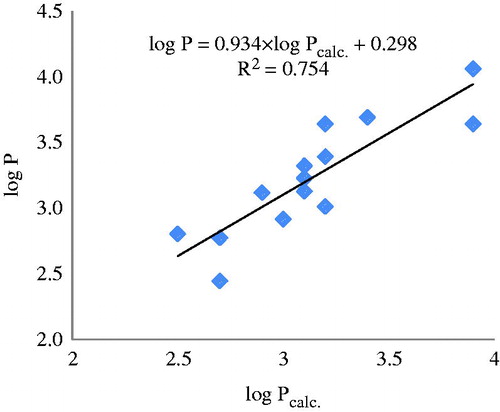
Table 1. Anthraquinone molecular structures and their log P (taken from PubChem).
Table 2. List of ligands showing their molecular weight and formula, hydrogen bond acceptors, hydrogen bond donors and torsions.
Figure 2. Glycogen synthase kinase-3 beta receptor, PDB Entry ID: 3Q3B, obtained from RCBS Protein data bank.
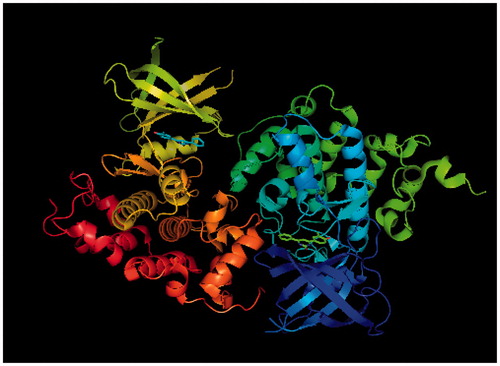
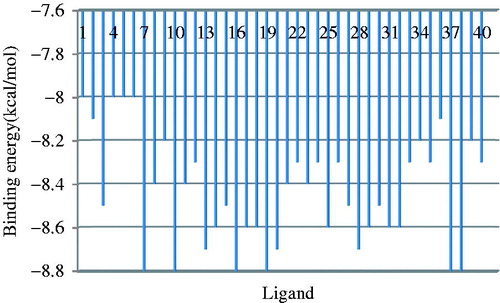
Table 3. Final lamarckian genetic algorithm docked state – binding energy for nine ligand conformations.
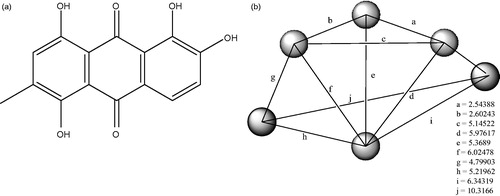
Figure 4. (a): Pharmacophore model for the receptor glycogen synthase kinase-3 beta. (b): Selected data on the pharmacophore model of anthraquinone/3Q3B protein interaction.