Figures & data
Scheme 1. Synthesis of inhibitor 35. Conditions and reagents (a) 1 equiv Boc-Ala-OH, 1 equiv BOP, 3 equiv DIPEA; (b) TFA, 1 h room temperature; (c) 1.15 equiv Boc2O in dioxane and water, pH ∼ 8.5 adjusted with 1 N NaOH; (d) 1 equiv BOP, 3 equiv DIPEA; (e) TFA, 1 h, room temperature, preparative HPLC.
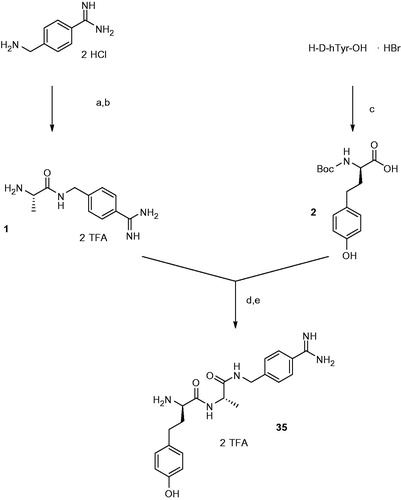
Figure 2. Analytical data and abbreviations of prepared racemic homo amino-acids. Their determined mass corresponds to the (M + H)+ ion, the HPLC analysis started at 1% solvent B.
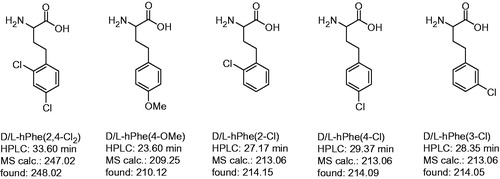
Table 1. Data collection and refinement statistics for the trypsin/inhibitor 35 complex.
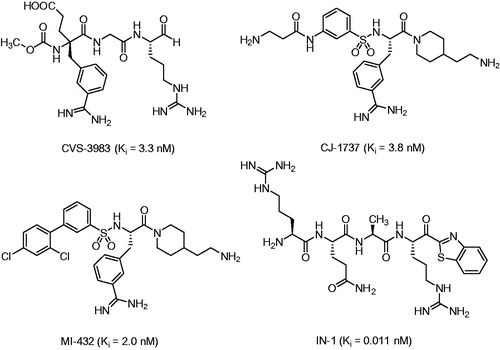
Table 2. Inhibition of matriptase, thrombin and fXa by benzylsulfonyl-protected inhibitors available from previous workCitation27,Citation49,Citation50.
Figure 3. Modeled binding mode of BAPA (inhibitor 5) in matriptase. The coordinates of matriptase were taken from the pdb entry 2gv6. The protease is shown with its solvent-accessible transparent surface in gray. The inhibitor is displayed as stick model with carbon atoms in white, nitrogen in blue, oxygen in red, and sulfur in yellow. Selected residues of matriptase forming the S3/4 pocket (Trp215 at the bottom, Phe99 on the right, Gln175 on the left and Phe97 on the top), as well as residues involved in polar contacts to the inhibitor (Asp189, Ser190, Gly216, and Gly219) are labeled and shown as sticks with carbons in green. All colored figures were prepared using PyMOL v0.98 (DeLano Scientific, San Carlos, CA).
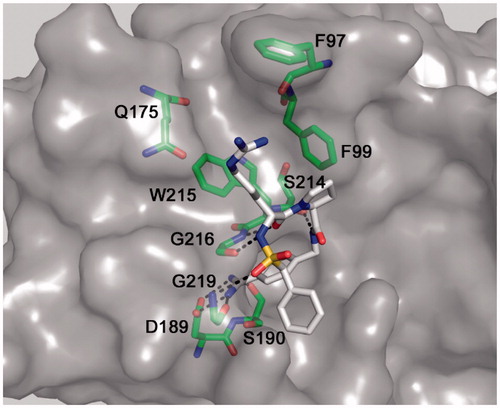
Table 3. Inhibition of matriptase, thrombin and factor Xa by inhibitors lacking a P4 residue.
Figure 4. Compound 35 in complex with trypsin and matriptase: (A) Crystal structure of compound 35, shown as sticks (carbon in green, nitrogen in blue, oxygen in red, and water molecules as red spheres) in complex with bovine trypsin presented with its backbone in beige (4MTB.pdb). Selected trypsin residues are shown as sticks with light pink carbon atoms; (B) Model of compound 35 in the active site of matriptase (shown with gray surface and backbone as cartoon in green), obtained by superimposition of the 35/trypsin complex with the crystal structure of matriptase (2GV6), followed by an energy minimization of the d-homotyrosine side-chain using the program MOECitation48. All other inhibitor atoms and the matriptase residues were fixed, water molecules were deleted. A pdb file of the modeled matriptase/inhibitor 35 complex is provided as Supplementary material for download.
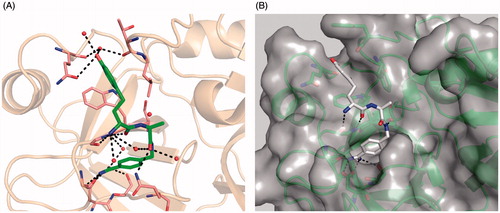
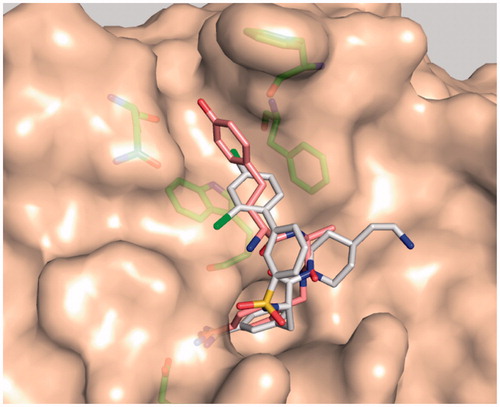
Figure 5. Model of matriptase in complex with inhibitor MI-432 (sticks with white carbon atoms, oxygen in red, nitrogen in blue, chlorine in green, structure shown in ) and the d-hTyr-derivative 35 (sticks with light pink carbons). The superpositions of both inhibitors reveals that their terminal phenyl rings occupy different positions in the distal S3/4 binding pocket. The matriptase residues Phe97, Phe99, Gln175, and Trp215, which shape the S3/4 pocket, and Asp189 are shown as sticks with green carbon atoms.