Figures & data
Figure 1. Structure of the HTD hit compound 1, of the optimized derivative 3i and general structure of inhibitors identified in this study.
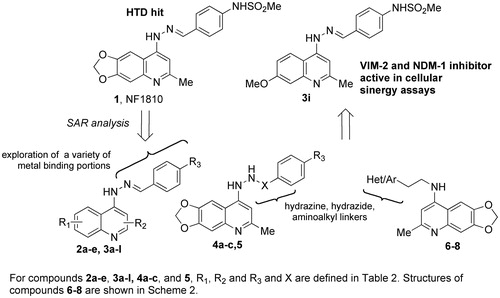
Table 1. Reaction conditions explored for hydrazone bond reduction for conversion of compound 2a to 4b (Scheme 1 and ).
Scheme 2. Synthetic procedure for the preparation of final compounds 6–8. Reagents and conditions: (a) TEA, EtOH, reflux, 76 h; (b) 2-aminoethanol, 150 °C, 15 min then 190 °C, 30 min (c) i. p-TsCl, pyridine, dry DCM, 0–25 °C, then 50 °C, 4 h; ii. 15 or 16, DIPEA, dry DMSO, 70 °C, 20 h.
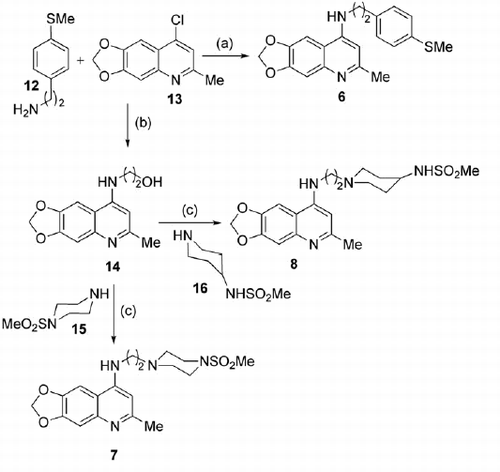
Table 2. Ki values for VIM-2 and NDM-1 for compounds, 1, 2a–e, 3a–l, 4a–c and 5–8.
Figure 2. Docked pose of 1 (sticks) into VIM-2 binding site (A), NDM-1 (B) and IMP-1 (C) as found by HTD protocol. Regarding IMP-1, two representative clusters were reported: in light sticks the docked pose of the first cluster, in dark sticks the docked pose of the second cluster. The contacts established by 1 into the active sites of the enzymes are represented by dotted lines. Metal ions were represented by spheres. The numbering of NDM-1 is referred to Uniprot KB protein sequence F6IAY7, while the numbering of IMP-1 is referred to Uniprot KB protein sequence P52699. The contacts established by 1 into the active sites of the enzymes are represented by dotted lines. Metal ions were represented by spheres. The numbering of NDM-1 is referred to deposited Uniprot KB protein sequence F6IAY7. Nonpolar hydrogens were omitted for the sake of clarity. The pictures were generated by PyMOL (The PyMOL Molecular Graphics System, v1.6-alpha; Schrodinger LLC, New York, 2013).
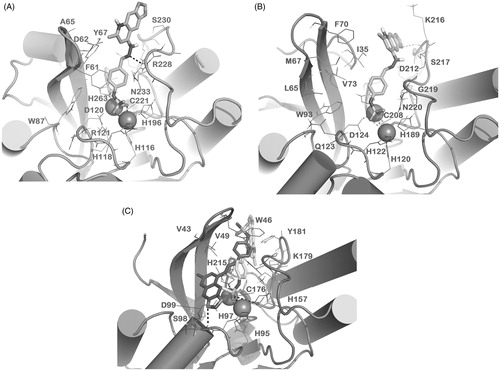
Figure 3. Docked pose of 3b into VIM-2 binding site (A) and NDM-1 (B) as found by docking protocol. The contacts established by the mentioned compounds into the active sites of the enzymes are represented by dotted lines. Metal ions were represented by spheres. Nonpolar hydrogens were omitted for the sake of clarity. For 3b in panel A, the clusters are as follows: cluster 1, ChemScore 38.86 and ΔGbind −61.74 kcal/mol; cluster 2, ChemScore 31.79 and ΔGbind −53.29 kcal/mol; cluster 3, ChemScore 27.11 and ΔGbind −40.73 kcal/mol). The pictures were generated using the PyMOL software.
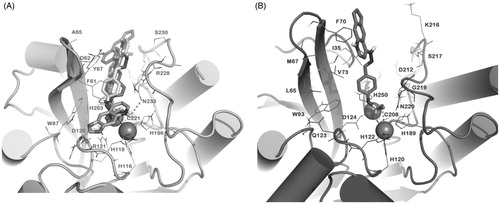
Figure 4. Docked pose of 3i (sticks) into VIM-2 binding site (A) and NDM-1 (B) as found by HTD protocol. The contacts established by 3i into the active sites of the enzymes are represented by dotted lines. Metal ions were represented by spheres. The numbering of NDM-1 is referred to deposited protein sequence (Uniprot KB code F6IAY7). Nonpolar hydrogens were omitted for the sake of clarity. The pictures were generated by PyMOL (The PyMOL Molecular Graphics System, v1.6-alpha; Schrodinger LLC, New York, 2013).
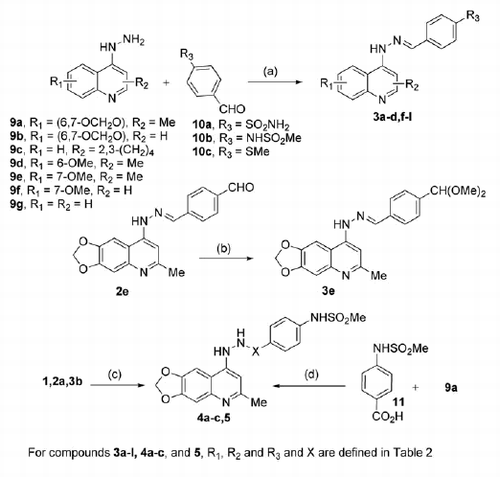
Scheme 1. Synthetic procedure for the preparation of final compounds 3a–l, 4a–c, and 5. Reagents and conditions: (a) EtOH, reflux, 2–16 h; (b) HBF4, MeOH, 25 °C, 2 min; (c) Et3SiH, TFA, 25 °C, 4–6 h; (d) DCC, HOBt, dry DMF, 25 °C, 12 h.