Figures & data
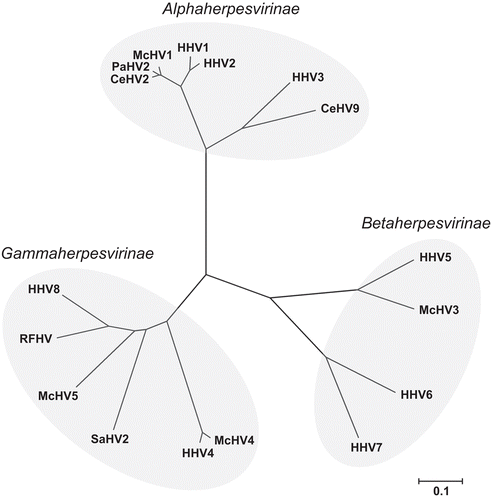
Figure 1. Schematic diagram of a herpesvirus demonstrating the major structural components including the genomic DNA, virus capsids, tegument, envelope, and envelope glycoproteins.
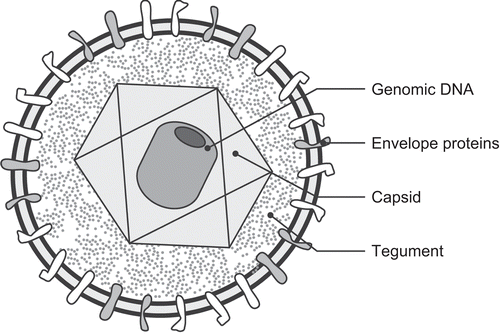
Table 1. Some primate viruses within the order Herpesvirales (Davison et al., Citation2009).
Figure 2. Evolutionary relationships of 17 common primate herpesviruses were inferred from the amino acid sequence of the DNA polymerase protein using the Neighbor-Joining method (Saitou and Nei, Citation1987). The tree is drawn to scale with branch lengths in units of number of amino acid substitutions per site used to infer the phylogenetic tree and evolutionary distances were computed using the Poisson correction method (Zuckerkandl and Pauling, Citation1965). Positions containing gaps and missing data were eliminated from the dataset resulting in a total of 941 positions in the final dataset. Phylogenetic analyses were conducted using MEGA4 (Tamura et al., Citation2007).