Figures & data
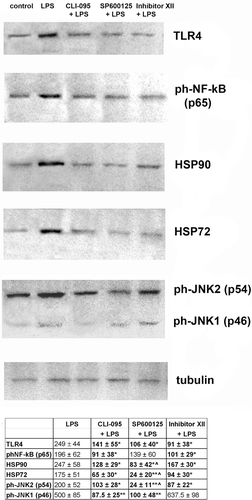
Figure 1. TLR4 signaling pathway and inhibitors of TLR4 and TLR4-related pathways used in the study. CD14, LBP, and TLR4 are part of the LPS recognition/response pathway. The most extensively studied signaling pathways activated by TLR4 in response to LPS require adaptor proteins including myeloid differentiation factor 88 (MyD88), MyD88 adaptor-like protein (Mal or, alternatively, TIRAP), TIR-containing adapter molecule (TRIF), and TRIF-related adaptor molecule (TRAM) (Krishnan et al., Citation2007). The differences in induction of MyD88-dependent and -independent signaling by endotoxin suggests that the initial interaction of an endotoxin with the extracellular domain of the TLR4 complex results in changes that lead to the recruitment of different adaptor proteins (Zughaier et al., Citation2005). MyD88 activation involves NF-κB and AP-1 signaling cascades, resulting in inflammatory cytokine over-production. The TLR4-mediated MyD88 independent pathway leads to dimerization and activation of IRF3 and IFNβ expression. It follows that the TLR4 signaling pathway leading to LPS-mediated NF-κB, AP-1, and IRF3 activation constitutes an important therapeutic target for sepsis therapy. Recognition of LPS and subsequent signaling pathways are not determined by one single receptor but through the orchestration of multiple receptors that are concentrated at so-called lipid rafts. For instance, in monocytes and macrophages several LPS signaling molecules, namely CD14, HSP70, and HSP90, are constitutively present at lipid rafts, while others, such as TLR4, CXCR4, and MyD88, are recruited upon LPS stimulation. It is known that LPS-mediated activation of the transcription factors NF-κB and AP-1 requires phosphorylation of IKK and JNK kinase, respectively. JNK subsequently phosphorylates AP-1, inducing its transactivation, whereas activated IKK phosphorylates the family of I-κB proteins, targeting them for degradation via the ubiquitin proteasome pathway (Medvedev et al., Citation2000; Haddad and Abdel-Karim, Citation2011). The early MyD88-dependent response to LPS also induces an early activation of IRF3 and induction of IFNκ. The MyD88-independent pathway is responsible for the delayed activation of NF-κB and IRF3. Intriguingly, there is a sequence of data indicating co-operation between the NF-κB and IRF3 pathways. Down-stream NF-κB, IRF3, and MAPK activation can result in intracellular over-production of pro-inflammatory cytokines, including TNFκ, IFNγ, IL-1, and IL-6, as well as reactive oxygen species, including nitric oxide (NO) (Akira et al., Citation2006; Novoselova et al., Citation2006; O’Neill et al., Citation2009). CLI-095 was used for specific inhibition of the TLR4 receptor protein, SP600125 for that of JNK 1/2/3 (which blocks the SAPK/JNK signaling pathway), and IKK Inhibitor XII for inhibition of IKKα/β/ϵ (which leads to NF-κB-pathway blockade).
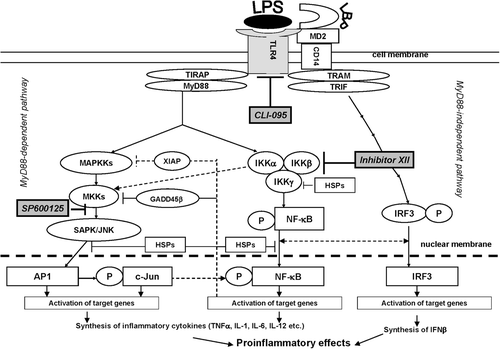
Figure 2. The effect of inhibitors on production of cytokines and NO by RAW 264.7 cells treated with LPS. RAW 264.7 cells from one separate passage were pooled, and aliquots of 1 × 106 cells (cultivated for 24 h) were used for each sample. NO was measured in supernatants, and cytokines were estimated using cellular lysates. Lysates of the cells were obtained by three cycles of freezing–refreezing, and cytokine concentrations in each sample were determined using ELISA. Results are expressed as the mean and standard error from four independent experiments that were performed separately with different passages; the measurements in separate experiments were made for each individual cellular sample (six repetitions). The effectiveness of inhibitors in comparison with the most effective, i.e. the IKK Inhibitor XII, is shown using upper staples. Value is significantly different from control, *p < 0.05, **p < 0.01; from LPS, #p < 0.05, ##p < 0.01; or from IKK Inhibitor XII, ^p < 0.05, ^^p < 0.01.
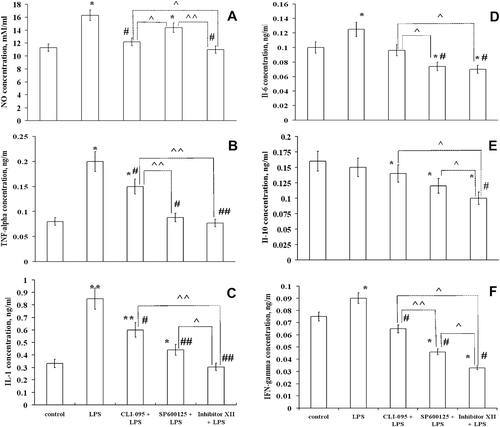
Figure 3. Inhibitors of cellular signaling pathways decrease expression of some of the proteins involved in LPS-induced response in RAW 264.7 cells. Expression of proteins was measured in cells lysed by boiling in Laemmli sample buffer. Equal amounts of protein were analyzed by Western blot analysis using corresponding antibodies. The values shown at the bottom in the Table were calculated as relative units and are the results of tested protein blots densitometry by program QAPA from four independent experiments, values are expressed as a percentage of control. Control values of the left blots were in all cases assumed to be 100%; tubulin is shown on a parallel blotting panel. Value is significantly different from LPS (*p < 0.05, **p < 0.01) or from IKK Inhibitor XII (^p < 0.05).