Figures & data
Figure 1. The GNPA protocol. The assay used MMP probes functionalized with monoclonal antibodies that recognize and bind target protein (i.e. ricin toxin). The ricin toxin was then functionalized with the nanoparticle (NP) probe that has been modified with ricin A chain antigen-specific antibodies and single-stranded signal DNAs. After being magnetically separated, the immune-complex containing the single-stranded signal DNA was characterized by PCR and by real-time PCR.
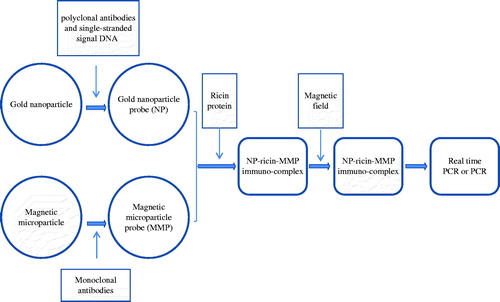
Figure 2. Specificity of GNPA using PCR detection. Signal DNA bound on GNP probe was analyzed using PCR. (Lanes a and b) Ricin A chain of two duplicates. (Lane c) Camphorin protein. (Lane d) Ebulitin protein. (Lane e) Blank control. (Lane m): DL2000 DNA marker.
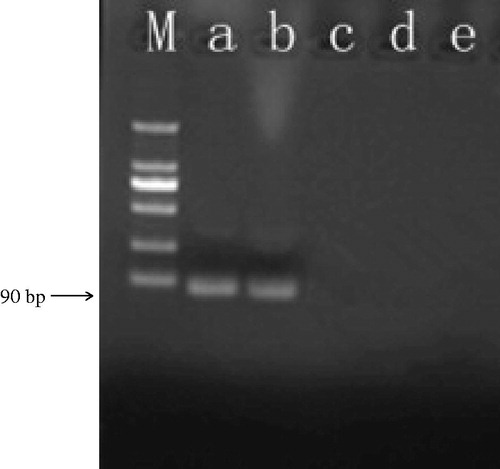
Figure 3. Specificity of the GNPA using real-time PCR detection. Signal DNA bound on GNP was analyzed using fluorescence real-time PCR. Real-time PCR results from: (Curves A, B) ricin A chain of two duplicates; (Curves C, D) camphorin and ebulitin protein; and (Curve E) blank control.
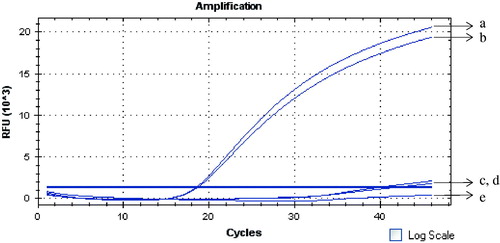
Figure 4. Sensitivity of the GNPA based on real-time PCR for the ricin A chain detection. Ten-fold dilutions of ricin A chain were subjected to the GNPA. Amplification plot of the signal DNA bound on GNP probe detected with real-time PCR showed that over 10−2 fg ricin A chain/ml were detected by the GNPA. (Curves A–F) amplification plots of 102, 101, 100, 10−1, 10−2, and 10−3 fg/ml ricin A chain detected by the GNPA. (Curve G) blank control.
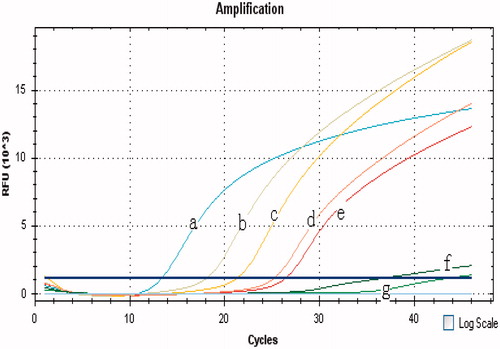
Table 1. Reproducibility of the GNPA for detection of ricin A chain.
