Figures & data
Figure 1. Curcumin induces cell surface expression of granule markers CD35, CD63, and CD66b in human PMN. Freshly isolated human PMN (10 × 106 cells/ml in complete RPMI 1640) were incubated with buffer (Ctrl), 10 μM curcumin (CUR10), 50 μM curcumin (CUR50), 10−9 M fMLP, or a mixture of CUR50 + fMLP for 30 min. The cell surface expression of (A) CD35, (B) CD63, and (C) CD66b was then assessed by flow cytometry. Results are in terms of the general mean (Gmean) of fluorescence expressed; shown are the mean ± SEM (n = 5) of these values. *p < 0.05 versus Ctrl; **p < 0.05 versus fMLP. Iso, isotypic control for the assay; ns = not significant.
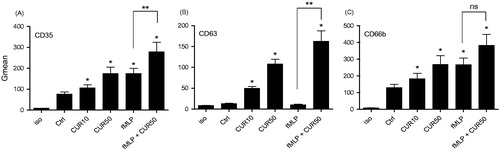
Figure 2. Curcumin increases MMP-9 expression and gelatinase activity in extracellular milieu. Freshly isolated human PMN (10 × 106 cells/ml in complete RPMI 1640) were stimulated for 30 min with buffer (Ctrl), 10 μM curcumin (CUR10), 50 μM curcumin (CUR50), 10−9 M fMLP, or a mixture of CUR50 + fMLP; supernatants were then harvested and used to perform (A) Western blots for analysis of MMP-9 protein expression or (B) zymography assays for determining enzymatic activity. (A, B) One representative experiments is shown (of six). the three gelatinase enzymatic activities (A, B, C) were similarly affected in all test conditions.
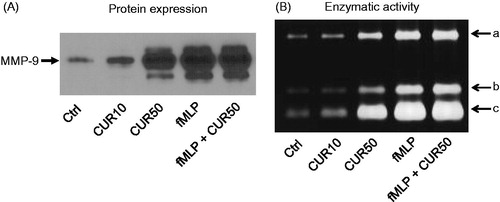
Figure 3. Curcumin does not further increase fMLP-induced p38 MAPK in human PMN and does not increase MMP-9 protein expression via p38 MAPK-dependent mechanism. (A) Cells (10 × 106 cells/ml complete RPMI 1640) were stimulated for 30 min with buffer (Ctrl), 10 μM curcumin (CUR10), 50 μM curcumin (CUR50), 10−9 M fMLP, or a mixture of CUR50 + fMLP; p38 MAPK activation was assessed by Western blot analysis. (B) Cells were treated as above but pre-incubated with diluent or 2 μM of p38 MAPK inhibitor (p38i) for 30 min; MMP-9 protein expression was studied by Western blot analysis. (A) Results from one representative experiment (of three). Equivalent loading was evaluated by expression of un-phosphorylated p38. (B) Densitometric analysis is plotted in a bar graph (integrated density value) to quantify MMP-9 protein expression (mean ± SEM, n = 3). Bottom: results from one representative experiment.
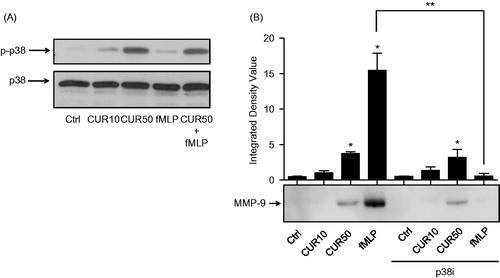
Figure 4. Ability of curcumin to increase gelatinase activity is unaffected by p38 MAPK inhibitor. PMN (10 × 106 cells/ml complete RPMI 1640) were pre-incubated with diluent or 2 μM of p38 MAPK inhibitor (p38i) for 30 min, and then treated for 30 min with buffer (Ctrl), 10 μM curcumin (CUR10), 50 μM curcumin (CUR50), or 10−9 M fMLP; supernatants were then harvested and underwent zymography assays. (A) Results from one representative experiment showing the three gelatinase activities (A, B, C). (D) Densitometric analysis of the three detected gelatinase activities were added together as each was similarly affected. Results are mean ± SEM (n ≥ 3). *p < 0.05 versus Ctrl; **p < 0.05 fLMP without p38i versus fMLP with p38i.
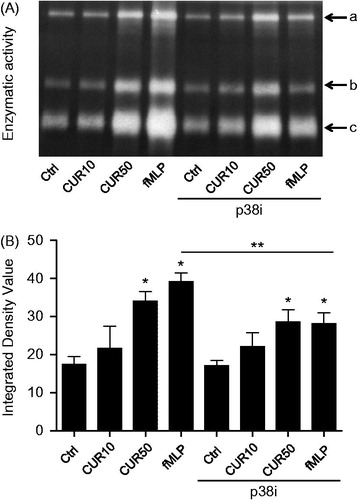
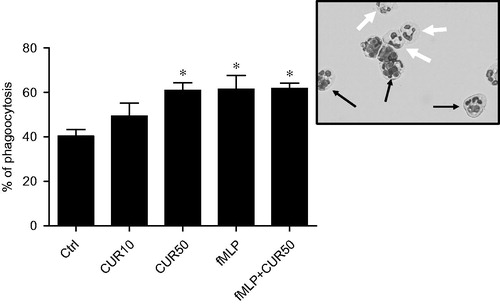
Figure 5. Curcumin enhanced PMN phagocytosis. Freshly isolated human PMN (10 × 106 cells/ml in complete RPMI 1640) were stimulated for 30 min with buffer (Ctrl), 10 μM curcumin (CUR10), 50 μM curcumin (CUR50), 10−9 M fMLP, or a mixture of CUR50 + fMLP. After incubating 106 PMN/treatment each with 5 × 106 opsonized SRBC for 30 min at 37 °C, the extent of phagocytic activity among the cells was assessed by counting the number of PMN placed on a slide that had ingested at least one opsonized SRBC. A minimum of five different fields (corresponding to ∼200 PMN/slide) was randomly assessed; each slide was analyzed in duplicate. Results are expressed as mean ± SEM (n ≥ 3). *p < 0.05 versus control. Inset: a typical PMN that did (thin black arrows) or did not (large black arrows) ingest an SRBC. Magnification = 400×.