Figures & data
Figure 1. Detection of the site of insertion by fluorescent in situ hybridization. Metaphase chromosomes were isolated from homozygous transgenic mouse lymphocytes and fixed onto slides. The site of insertion was detected using a fluorescently labeled cosmid clone hp3.1 (red and arrows). Individual chromosomes were identified by DAPI banding. FISH signals from 100 fields were observed under fluorescence. The hybridization signal was localized to mouse chromosome 19, region C3.
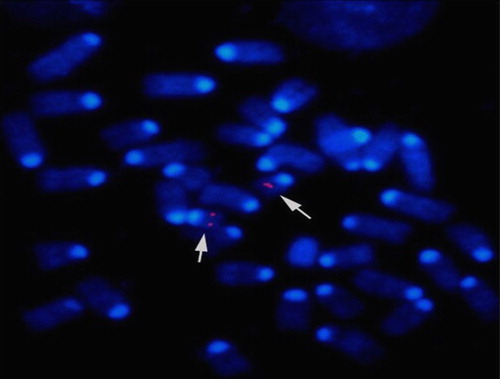
Table 1. Primers Used for Determination of Copy Number and Relative Expression Levels.
Figure 2. GENOMIC LOCATION OF INSERTION SITE. The cosmid DNA fragment encompassing the human protamine domain (chromosome 16; 11,349,856 - 11,390,141; red hatched box), including the PRM1, PRM2, and TNP2 genes flanked by boundary elements, was determined by terminal transferase dependent PCR to have inserted within cytological band C3 of mouse chromosome 19. Integration occurred within a L1Md T repeat element of the seventh intron of the cytochrome P450 2c38 gene.
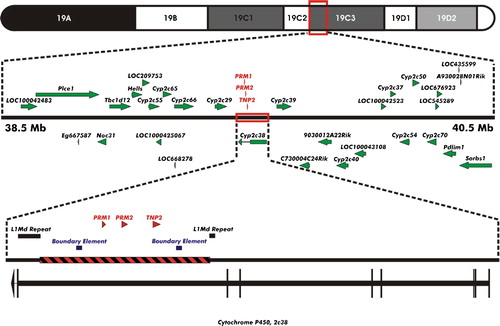
Figure 3. Relative levels of the endogenous and human protamine transcripts in transgenic mice. Transcript levels were assessed by qRT-PCR of total testis RNA. A). The relative levels of the endogenous protamine gene transcripts prm1, prm2, and tnp2 were not inhibited in transgenic animals harboring the human protamine construct compared to wild-type mouse. The level of β-actin provides a comparator of the variance among the animals. B) The relative levels of the human PRM1, PRM2, and TNP2 transcripts. Transgene expression was similar to that observed in human males. Values are medians +/− two standard deviations of triplicate reactions.
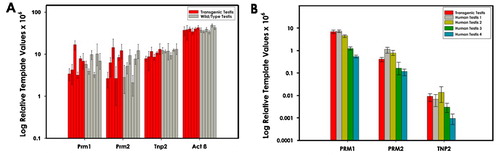
Table 2. Genomic Coordinates of the Regions of Interest Represented on the Tiling Array.
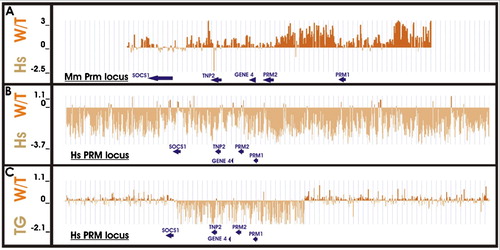
Figure 5. Interspecies comparative CGH array analysis of human, wild-type, and transgenic mouse genomic DNA. Competitive hybridization of wild-type mouse and human genomic DNA to probes targeting A) the mouse protamine domain (chromosome 16: 10,782,515 – 10,802,516; GRCh37) and B) the extended human protamine domain (chromosome 16: 11,312,500–11,452,413; GRCh37). Both sets of probes exhibit species specificity. C) Competitive hybridization of wild-type and homozygous transgenic mouse genomic DNA to probes targeting the extended human protamine domain confirms insertion of the complete transgenic locus. Elevated transgenic signal is directly representative of the inserted human DNA sequence (chromosome 16: 11,349,856–11,390,141; GRCh37); signal ratio corresponding to the regions flanking the protamine domain is ∼1:1. Y axis is Log2.