Figures & data
Figure 1. Effect of long-term hESC culture without exogenous FGF2. The pooled analysis revealed a significant up-regulation of six genes related to endothelial (FLT-1 and PTF1A-p48), neuronal cells (PAX6 and NR6A1), as well as pluripotency (POU5F1) and cancer related genes when up-regulated (EEF1A1). Relative quantity was calculated by ddCt method for each sample, from the mean of four replicates. Error bars: SEM: standard deviation of the mean, Significance: *P < 0.05, **P < 0.01
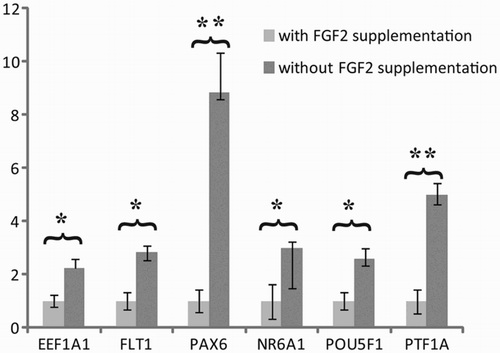
Figure 2. Stem (POU5F1, A) and germ cell (DDX4, B) markers pooled (A and B) and single evaluation (a-f) in three hESC lines after stimulation with ATRA for 7 days in vitro. No up- or down-regulation could be observed by Q-PCR gene expression analysis after supplementation with ATRA when all cell lines were pooled (A and B). Gene expression of germ cell markers in LRB017 showed a positive response to the supplementation with ATRA when compared to day 0. Day 0 is control hESCs that have been spontaneously differentiated on hFFs for 12 weeks and showed a significant higher expression of POU5F1 and DDX4 genes compared to hFF alone. Relative quantity was calculated by ddCt method for each sample, from the mean of three replicates. Error bars: SD: standard deviation, Significance: ***P < 0.001
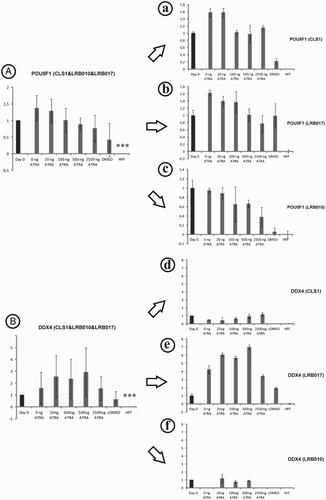
Figure 3. Western blot analysis of DDX4 and POU5F1 in LRB017 after stimulation with ATRA for 7 and 14 days. ATRA is all-trans retinoic acid dissolved in DMSO. Positive control for DDX4 is human adult testis and for POU5F1 is undifferentiated hESCs (LRB017). All markers analyzed DDX4, POU5F1, and GAPDH showed bands of expected size (75kDa, 43kDa, and 35kDa, respectively).
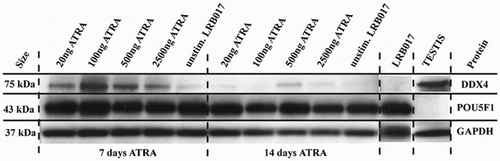
Figure 4. DDX4 expression at RNA level in 8 undifferentiated hESC lines and 3 hiPSC lines established in 5 different laboratories. DDX4 expression was detected at RNA level in all cell lines analyzed. Relative quantity was calculated by ddCt method for each sample from mean of three replicates.
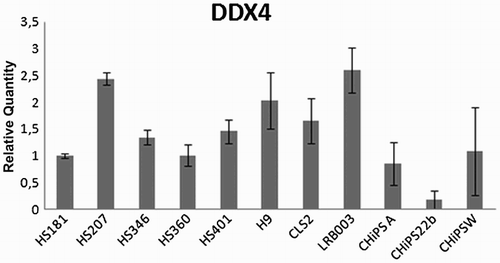
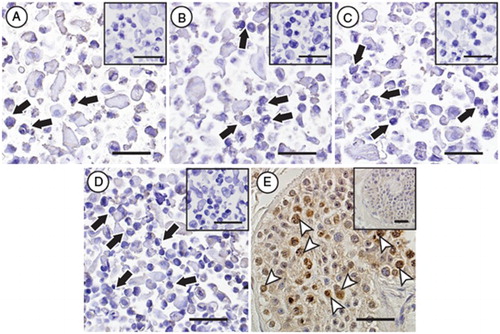
Figure 5. Immunocytochemical evaluation of DDX4 expression in hESC lines. No specific protein expression of DDX4 could be observed in any of the evaluated stem cells lines (black arrows, H9 (A), HS207 (B), HS360 (C) and HS401 (D)), whereas positive protein expression of DDX4 (white arrow heads) could be observed in male germ cells in cross-sections of a human testicular biopsy (E). Negative controls for the primary antibody for all samples are shown as small Figures (A-E). Scale bars: 50µm.