Figures & data
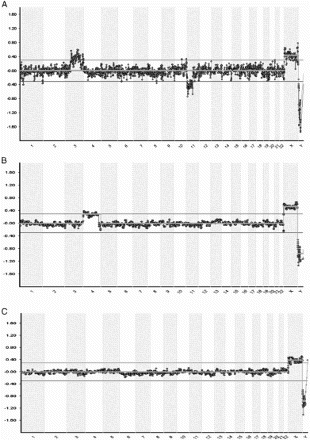
Figure 1. Indirect preimplantation genetic diagnosis (PGD) for spinal muscular atrophy. A) Location and distances to the SMN1 gene of the short tandem repeat microsatellite markers used. B) Family tree showing the relatives involved in the informativity study and PGD cycle. Both members of the couple are carriers of a deletion involving exons 7 and 8 in the SMN1 gene. The dotted line represents the paternal disease-bearing haplotype. The thick black line illustrates the maternal disease-bearing haplotype. C) Electropherograms of the PGD for spinal muscular atrophy. Examples for two of the embryos analyzed. The numbers represent fragment sizes in base pairs for every allele. Numbers in bold stand for the alleles associated to the maternal mutation; numbers in bold and italics correspond to the alleles associated to the paternal mutation. Cen: centromere; tel: telomere; Mb: megabases; ADO: allele drop-out.
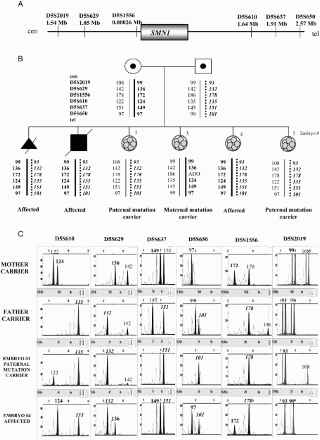
Figure 2. Direct preimplantation genetic diagnosis (PGD) for Huntington disease. A) Position and distances to the HTT gene of the short tandem repeat microsatellite markers involved in the informativity study and PGD cycle. B) Genealogic tree of the family taking part in the study and PGD results. The arrow designates the consultant affected patient. The thick black line illustrates the expansion-bearing haplotype. C) Electropherograms of the PGD for Huntington disease. Some examples of the embryos analyzed are displayed. The numbers represent fragment sizes (in base pairs) for every allele. Numbers corresponding to the disease-bearing allele are in bold. Cen: centromere; tel: telomere; Mb: megabases; ADO: allele drop-out.
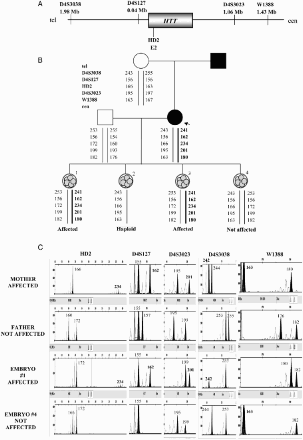
Figure 3. Direct PGD for cystic fibrosis. A) Location and distances to the CFTR gene of the short tandem repeat microsatellite markers used in the preclinical study and preimplantation genetic diagnosis (PGD) cycle. B) Pedigree of the family involved in the study and PGD results. The mother carries the mutation c.2988 + 1G > A in intron 16 of the CFTR gene. The father carries the mutation c.489 + 1G > T in intron 4 of the CFTR gene. The affected daughter carries both mutations in compound heterozygosis. These two positions were minisequenced in the embryos. The dotted line represents the paternal disease-bearing haplotype. The thick black line represents the maternal disease-bearing haplotype. C) Electropherograms of the PGD for cystic fibrosis. Some examples are shown for embryos 1 and 4. The numbers represent fragment sizes for every allele in base pairs. Numbers in bold represent the alleles associated to the maternal mutation; numbers in bold and italics correspond for the alleles associated to the paternal mutation. Cen: centromere; tel: telomere; Mb: megabases; G:guanine; A: adenine; T: timine.
Figure 4. Combined preimplantation genetic diagnosis (PGD) for Fanconi anemia combined with human leukocyte antigen (HLA) matching. A) Location of the short tandem repeat microsatellite marker D16S520 with respect to the FANCA gene on chromosome 16. B) Genealogic tree of the family involved in the preclinical study and PGD results. The arrow indicates the consultant patient. Haplotypes for the HLA region on chromosome 6 plus Fanconi anemia mutations positions on chromosome 16 were established. The discontinuous line and the fine line represent the HLA haplotypes from the mother and the father, respectively, that embryos should inherit in order to be HLA compatible with the affected son. The thick black line symbolizes the disease-bearing haplotype for the paternal mutation (c.3788_3790delTCT) and the dotted line denotes the disease-bearing haplotype for the maternal mutation (c.4130C > G). Mutation c.3788_3790delTCT was determined by fragment analysis and position c.4130 of the FANCA gene was minisequenced. The genetic status for every embryo obtained is differentiated between HLA compatibility and Fanconi anemia diagnosis. Cen: centromere; tel: telomere; Mb: megabases; C: cytosine; G:guanine.
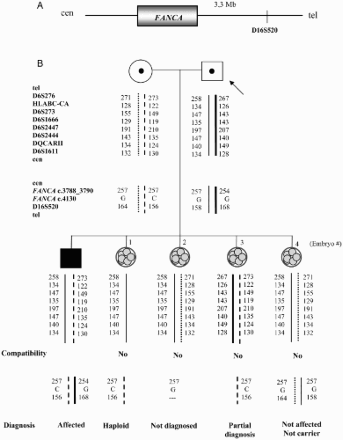
Figure 5. Example of blastomere array- comparative genomic hybridization charts. Chromosomal position is represented on the X-axis and the Log2 ratio Ch1/Ch2 (the ratio of red/green fluorescence intensity on each clone) on the Y-axis. A) Female blastomere with gain of chromosome 3(p14.2→qter) and loss of chromosome 11(q12.3→qter). The father carries a balanced translocation with chromosome formula: 46,XY;t(3;11)(p21;q13). B) Array profile showing a female cell with gain of chromosome 4. C) Normal female profile.