Figures & data
Figure 1. Blastocyst stained using the trophectoderm (TE) selective labeling method. A1) TE cells labeled with Alexa Fluor 488-conjugated wheat germ agglutinin (WGA). Arrowhead shows inner cell mass (ICM) location, where green fluorescence intensity is lower. A2) TE and ICM nuclei staining with propidium iodide. A3) merged A1 and A2 images.
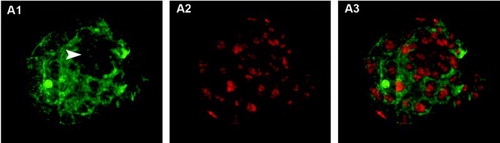
Figure 2. Blastocysts stained using trophectoderm (TE) permeabilization. Column 1 (A1, B1, C1) shows all blastocyst nuclei stained with Hoechst 33258 (blue). Column 2 (A2, B2, C2) shows TE nuclei stained with propidium iodide (PI, red). Column 3 (A3, B3, C3) shows merged images where TE nuclei are pink and inner cell mass (ICM) nuclei are blue. A1-A3) correctly stained blastocyst. Arrowhead points to the condensed chromatin of the second polar body and arrow indicates a dividing TE cell. B1-B3) incorrectly stained blastocyst showing poor PI staining, probably due to insufficient permeabilization of TE cells. C1-C3) incorrectly stained blastocyst showing PI staining in all nuclei, probably due to excessive cell permeabilization.
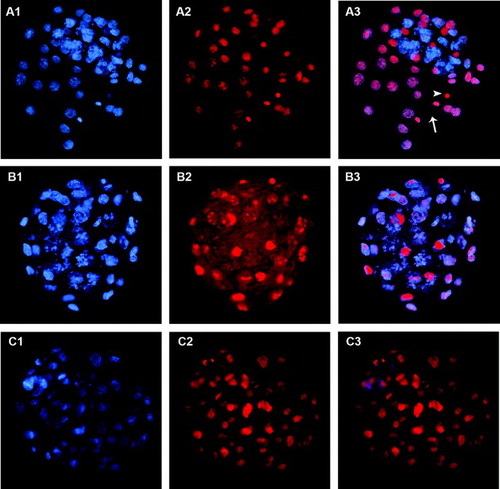
Figure 3. Blastocyst stained using the immunodetection method. A1) Hoechst 33258 staining of all nuclei. A2) Cdx2 staining of trophectoderm (TE cells). A3) Oct4 staining of inner cell mass (ICM cells). A4) merged A2 and A3 images. White arrowheads indicate four dividing TE cells where Cdx2 spreads over the whole cytoplasm. The yellow arrowhead indicates a nucleus coexpressing Cdx2 and Oct4.
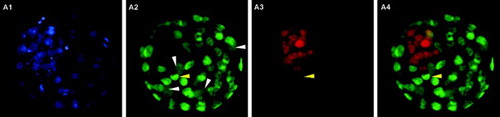
Table 1. Efficiencies of the TE permeabilization and immunodetection methods for the differential staining of mouse blastocysts.
Table 2. Blastocyst cell counts for the TE permeabilization and immunodetection methods.