Figures & data
Table 1. Purification steps of lipase from wheat germ.
Figure 3. FTIR graphs of (a) poly(HEMA), (b) poly(HEMA-MATrp) and (c) poly(HEMA-MATrp)-Cu(II) cryogels.
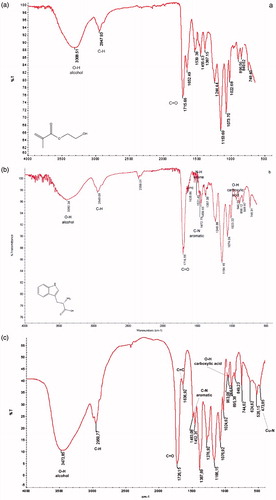
Figure 4. Effect of pH on adsorption of lipase onto poly(HEMA-MATrp) (Clipase: 1.5 mg/mL, interaction time: 40 min., temperature: 25 °C) and poly(HEMA-MATrp)-Cu(II) (Clipase: 2.0 mg/mL, interaction time: 30 min., temperature: 25 °C) cryogels.
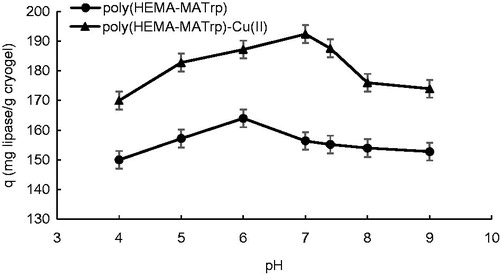
Figure 5. Effect of interaction time on adsorption of lipase onto poly(HEMA-MATrp) (Clipase: 1.5 mg/mL, pH: 6.0, temperature: 25 °C) and poly(HEMA-MATrp)-Cu(II) (Clipase: 2.0 mg/mL, pH: 7.0, temperature: 25 °C) cryogels.
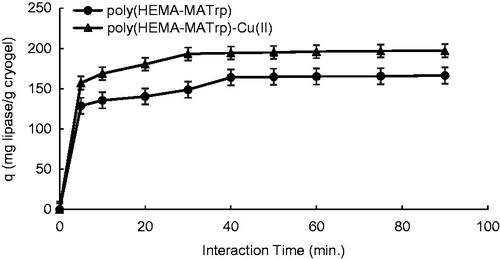
Figure 6. Effect of temperature on adsorption of lipase onto poly(HEMA-MATrp) (pH: 6.0, interaction time: 40 min., CLipase: 1.5 mg/mL) and poly(HEMA-MATrp)-Cu(II) (pH: 7.0, interaction time: 30 min., CLipase: 2.0 mg/mL) cryogels.
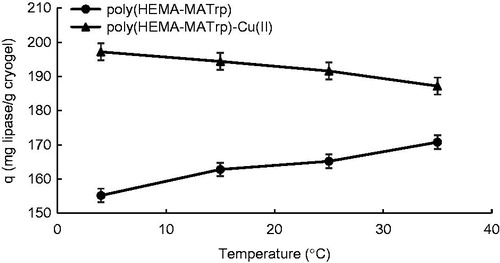
Figure 7. Effect of ionic strength on adsorption of lipase onto poly(HEMA-MATrp) (pH: 6.0, interaction time: 40 min., CLipase: 1.5 mg/mL, temperature: 25 °C) and poly(HEMA-MATrp)-Cu(II) (pH: 7.0, interaction time: 30 min., CLipase: 2.0 mg/mL, temperature: 25 °C) cryogels.
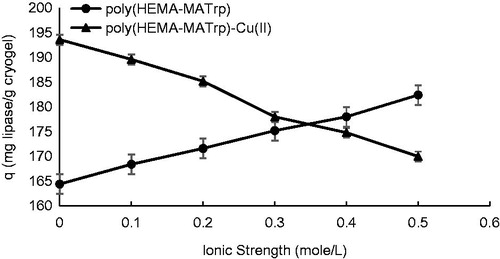
Figure 8. SDS-PAGE of desorbed lipase obtained from Candida cylindracea. Lane 1. Lipase, lysozyme, and hemoglobin marker, Lane 2: Lipase marker, Lane 3: Initial [(Before adsorption for poly(HEMA-MATrp)], Lane 4: Final [(After adsorption for poly(HEMA-MATrp)], Lane 5: Desorbed sample [(After dialysis for poly(HEMA-MATrp)], Lane 6: Initial [(Before adsorption for poly(HEMA-MATrp)-Cu(II)], Lane 7: Final [(After adsorption for poly(HEMA-MATrp)-Cu(II)], Lane 8: Desorbed sample [(After dialysis for poly(HEMA-MATrp)-Cu(II)].
![Figure 8. SDS-PAGE of desorbed lipase obtained from Candida cylindracea. Lane 1. Lipase, lysozyme, and hemoglobin marker, Lane 2: Lipase marker, Lane 3: Initial [(Before adsorption for poly(HEMA-MATrp)], Lane 4: Final [(After adsorption for poly(HEMA-MATrp)], Lane 5: Desorbed sample [(After dialysis for poly(HEMA-MATrp)], Lane 6: Initial [(Before adsorption for poly(HEMA-MATrp)-Cu(II)], Lane 7: Final [(After adsorption for poly(HEMA-MATrp)-Cu(II)], Lane 8: Desorbed sample [(After dialysis for poly(HEMA-MATrp)-Cu(II)].](/cms/asset/5cdcf2fa-fe0b-4192-b43e-54075ad71219/ianb_a_1161642_f0008_c.jpg)
Table 2. Parameters for adsorption isotherms of poly(HEMA-MATrp) and poly(HEMA-MATrp)-Cu(II).