Figures & data
Fig. 1 The old and new tularemia foci in Bulgaria. A. The genetic distance between strains L2 and 81 (orange dots) comprised five SNPs in 33 years. B. The genetic distance between strain 94 from USSR and strain 19c from Srebarna (yellow dots) comprised two SNPs in 5 years. C. There was an accumulation of >20 SNPs between the Srebarna 19 from the old focus and L1, MN, and MMM in the new focus (red dots). D. The first Bulgarian genome in the B.4 basal clade was represented by strain B1 isolated in Breznik in 1999 (black dot).
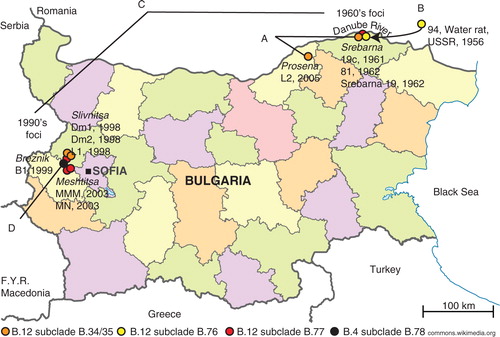
Table 1 Francisella tularensis strains investigated in the study
Table 2 Genome data for 10 Bulgarian strains and one archive strain sequenced in this study
Table 3 Four new assays B.76−B.79 for Bulgarian clades. Existing assays using modified primer sequences from previous publications
Fig. 2 Phylogeny. Whole genome phylogeny of F. tularensis subsp. holarctica based on SNPs in an alignment of nine published genomes (strain names are indicated) showing the placement of the Bulgarian strains in a global phylogenetic framework. Bulgarian outbreak strains and the non-outbreak strain 94 sequenced in this study are indicated in bold text. B.4, B.6, B.12, and B.16 indicate the four major canSNP clades of the subspecies. B.20 and B.23 indicate two B.12 subclades that included Bulgarian strains The tree was mid-point rooted in FSC022. The total length of the root branch was 3,317 base differences.
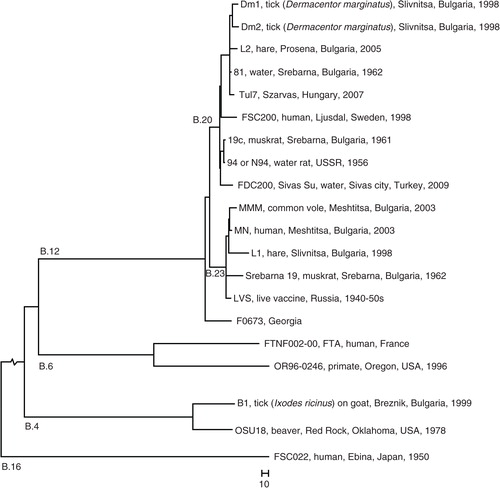
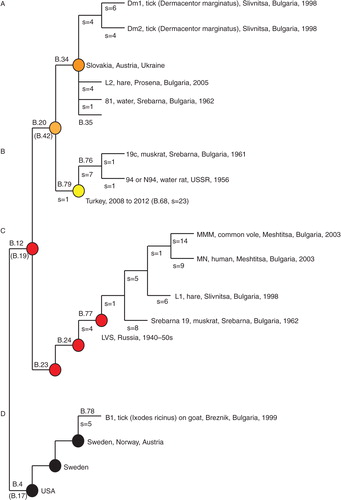
Fig. 3 The new Bulgarian genomes placed in the phylogenetic framework of canSNPs. Four assays B.76 − B.79 were developed in this study to target subclades, where strains from Bulgaria were members. B.4, B.12, B.23, B.24, B.34, and B.35 have been previously published (Citation5, Citation6) (Citation20). Alternative canSNPs (B.17, B.19, and B.42) are indicated in gray below the first published canSNP in indicated branch (Citation6, Citation19). s=the numbers of canSNPs on the indicated branch.