Figures & data
Fig. 1 Snapshot of EVpedia homepage (http://evpedia.info). General introduction of EVs and statistics of EVpedia are provided in the “Home” menu.
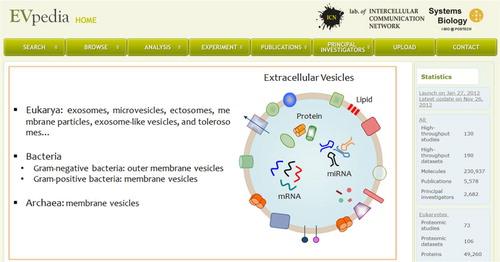
Table I Statistics of EVpedia, Exocarta, and Vesiclepedia
Table II Analytic tools in EVpedia, Exocarta, and Vesiclepedia
Fig. 2 Browse function of EVpedia. With “Browse” menu, the protein list from the individual dataset or all the datasets can be browsed. In this figure, we browsed the vesicular proteins of the urine from normal donors (Citation20).
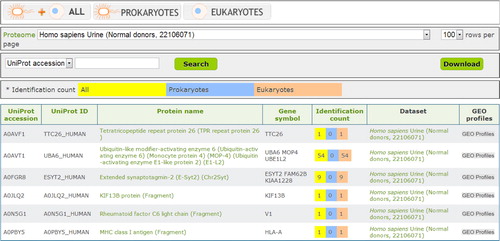
Fig. 3 Gene Ontology enrichment and network analyses. All analyses in this figure involved mRNA transcriptome of EVs derived from Homo sapiens mast cell HMC-1 (Citation21). In the “Analysis – Gene Ontology enrichment analysis” menu (a), by defining the species from which the analyzed list of proteins originates, the type of Gene Ontology terms (i.e. biological process, molecular function, and cellular component), and the cut-off of the enrichment p-value, the Gene Ontology enrichment analysis can be performed. In “Analysis – Network analysis” menu (b), by defining the species to which the functional interactome data belong, the number of additional nodes, and confidence of the interactome data, one can perform the network analysis of a protein list in EVpedia.
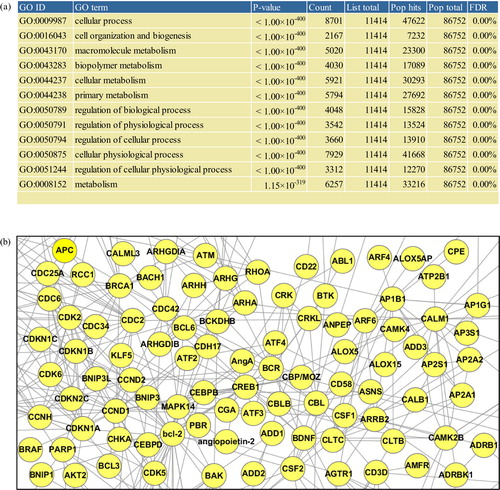
Fig. 4 Set analysis of EVpedia. In “Analysis – Set analysis” menu, the dataset of the “proteome+transcriptome (mRNA)”, “transcriptome (miRNA)”, and “lipidome” can be chosen for set analysis. Especially for the datasets of “proteome+transcriptome (mRNA)”, information from ortholog identification is used to build the Venn diagram (a). For example, the Venn diagram is drawn by comparing the ortholog clusters of vesicular proteomes from Homo sapiens SW480 and SW620 colorectal cancer cells (Citation20). The functional network of SW480-specific vesicular proteins can be drawn (b).
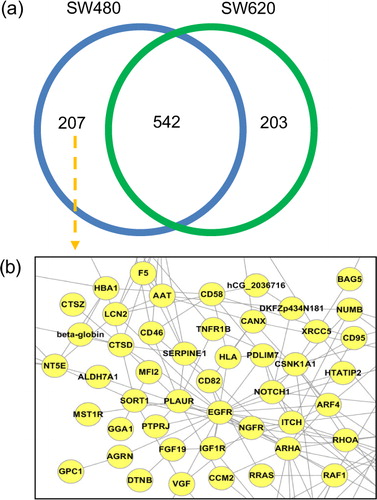
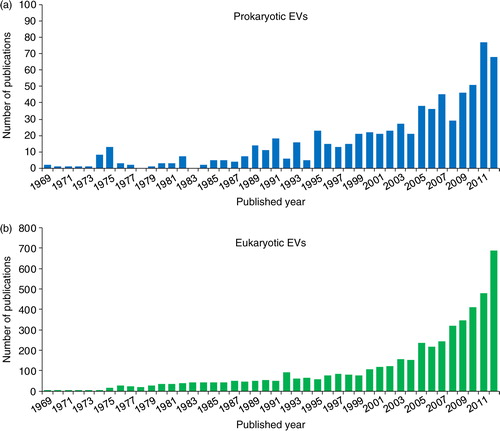
Fig. 5 Publications in EVpedia. In “Publications” menu, one can browse papers related to prokaryotic (a) and eukaryotic (b) EVs. The bar graph shows the number of EV publications for each year. One can search the papers with a keyword in the type of “category”, “bibliography”, “title”, “author”, and “title+abstract”.