Figures & data
Fig. 1. Effects of Porphyromonas gingivalis (Pg) on oral epithelial cells. (A) Pg W83, Streptococcus oralis 180B (So180B), or Pg+So180B were added to OKF6/TERT2 cells. Cocultures were allowed to incubate in 5% CO2 at 37°C for 16 hr and then examined by blinded investigators. (B) OKF6/TERT-2 cells were grown to confluence and used in the wound-healing assay. A linear streak was made in the cell monolayer. Pg, So180B, or Pg+So180B were added to the cultures at MOI = 100. After incubation for 2 hrs, the plates were washed to remove bacteria and fresh media was added. The cultures were incubated for 16 hrs and read by blinded investigators.
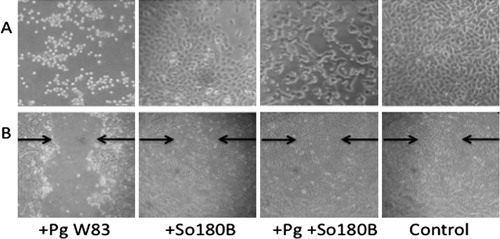
Table 1. Inhibition of Porphyromonas gingivalis (Pg) cytotoxicity by clinical strains
Table 2. Effects on Porphyromonas gingivalis (Pg) gingipain activity and production of acid and hydrogen peroxide by oral bacterial strain
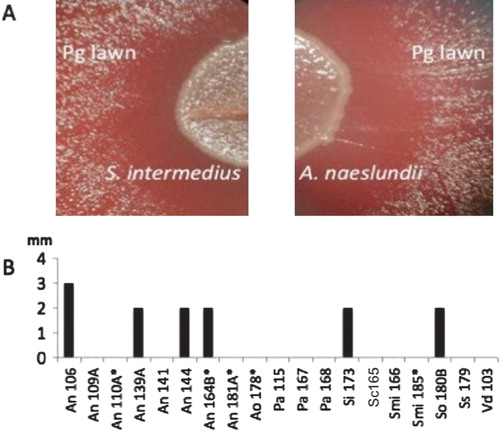
Fig. 2. Effect of oral bacterial isolates on Porphyromonas gingivalis (Pg) growth. Broth cultures of 19 clinical strains identified as inhibitory to Pg cytotoxicity were spotted on blood agar inoculated with a lawn of Pg and incubated anaerobically at 37°C for 48 hrs. The growth of Pg was inhibited near the colonies of 6 of 19 strains including (A) Streptococcus intermedius and Actinomyces naeslundii. (B) Growth inhibition was measured as the size of the clear zone (in mm) between the edge of the colony and the lawn of P. gingivalis. Bars represent the average of triplicate measurements confirmed in three independent experiments. Abbreviations used: An, Actinomyces naeslundii; Pa, Propionibacterium acnes; Si, Streptococcus intermedius; Smi, Streptococcus mitis; Sc, Streptococcus constellatus; So, Streptococcus oralis; Streptococcus sanguinis; Vd, Veillonella dispar. *less than 98% 16S rRNA sequence similarity to the closest type strain.