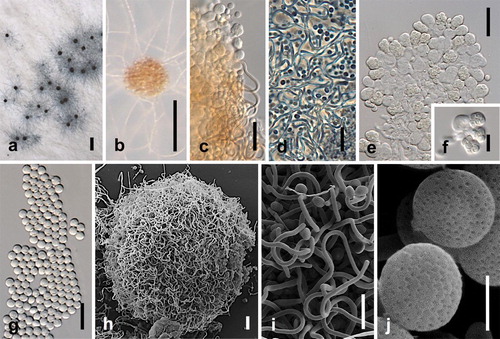
Figures & data
Fig. 1. Growth on different media and temperatures. A. Growth on MEA in the dark at 5 C intervals 5–40 C after 7 d. B. Growth on eight different media in the dark at 25 C after 14 d. Bars = standard errors.
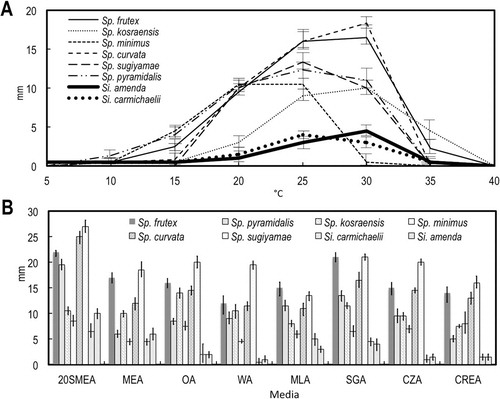
Fig. 2. Colonies of six new species of Spiromastix and two new species of Sigleria on four media after 14 d in the dark at 25 C. In each pair of photographs the top is the obverse and the bottom is the reverse.
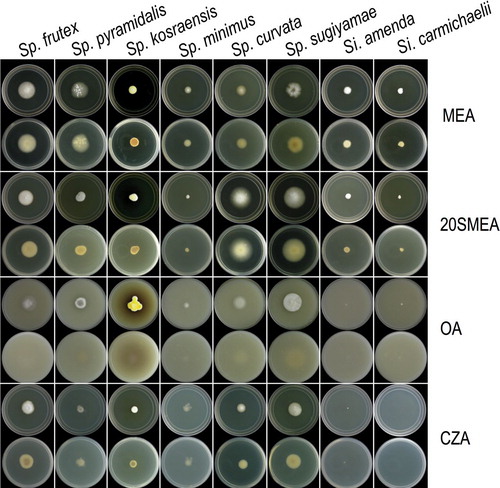
Fig. 3. Graphs of 95% confidence intervals of lengths (open shapes) and widths (gray shapes) of conidia, with means indicated by the constriction. A. Terminal and single lateral conidia. B. Arthroconidia. Bars = standard errors.
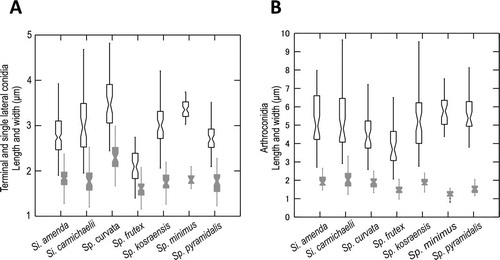
Fig. 4. Bayesian tree based on 18S + 28S sequences showing the relationship between the new genus Sigleria and new species of Spiromastix, their placement in the family Spiromastigaceae among other taxa of Onygenales and Eurotiomycetes. Clades with 1.00 Bayesian inference posterior probabilities (BI-PP) and 100% maximum likelihood bootstrap branch support (ML-BS) are indicated by thick black lines. Clades with > 0.95 BPP and 80% ML-BS are indicated by thicker gray lines, with the corresponding support values indicated; the BI-PP value is to the left and the ML-BS to the right. Asterisks indicate Bayesian posterior probabilities of 1.00 or maximum likelihood bootstrap branch support of 100%. Dashes indicate support values lower than 0.96 BPP or 80% ML-BS. Some support values for terminal branches were removed to reduce clutter. GT indicates that the strain represents the type species of its genus and ET indicates ex-type cultures. GenBank accession numbers for all sequences are provided (Supplementary table I).
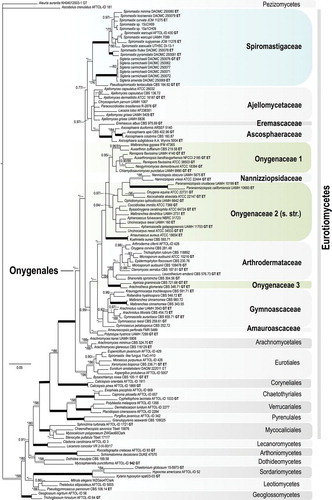
Fig. 5. Bayesian tree based on ITS sequences of the relationship between the new genus Sigleria and new species of Spiromastix, their placement in the family Spiromastigaceae among other families of Onygenales. Symbols and coding the same as for . GenBank accession numbers for all sequences are provided (Supplementary table I).
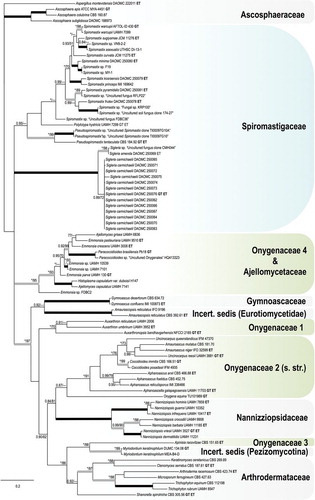
Fig. 6. Sigleria amenda. Ex-type culture on MEA. A. Sporodochial and aerial conidiophores. B–F. Conidiophores and conidia. G, H. Terminal and single lateral conidia and arthroconidia. I. arthroconidia. Bars: A = 500 μm, B–J = 10 μm.
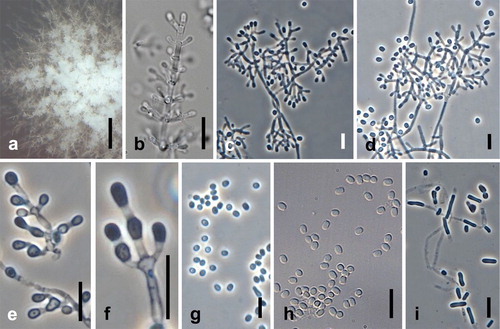
Fig. 7. Sigleria carmichaelii. Ex-type culture on MEA. A. Sporodochial and aerial conidiophores. B–E. Conidiophores and conidia. F–H Conidia. I, J. Chlamydospores. Bars: A = 500 μm, B–J = 10 μm.
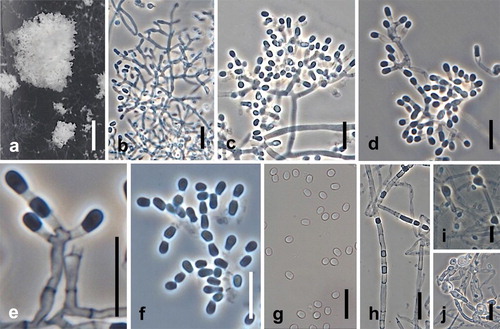
Fig. 8. Spiromastix curvata. Ex-type culture on MEA. A. Aerial conidiophores. B–D. Conidiophores and conidia. E–G. Terminal and single lateral conidia and arthroconidia. H. Chlamydospores. Bars: A = 500 μm, B–H = 10 μm.
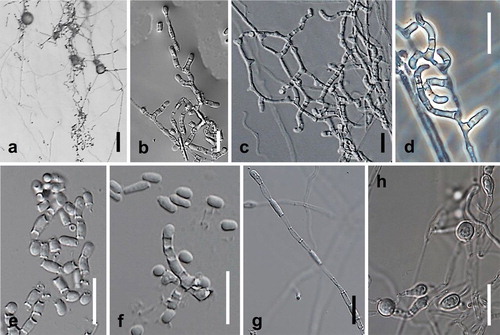
Fig. 9. Spiromastix frutex. Ex-type cultures on MEA. A. Aerial conidiophores. B, C. Conidiophores and conidia. D–G. Terminal and single lateral conidia and arthroconidia. Bars: A = 500 μm, B–G = 5 μm.
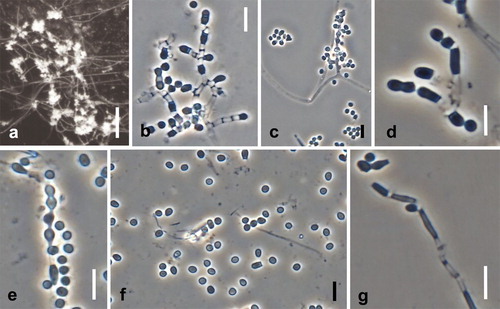
Fig. 10. Spiromastix kosraensis. Ex-type culture on MEA. A, B. Aerial conidiophores. C–G. Conidiophores and conidia. H, I. Terminal and single lateral conidia and arthroconidia. Bars: A = 50 μm, B = 100 μm, C–I = 10 μm.
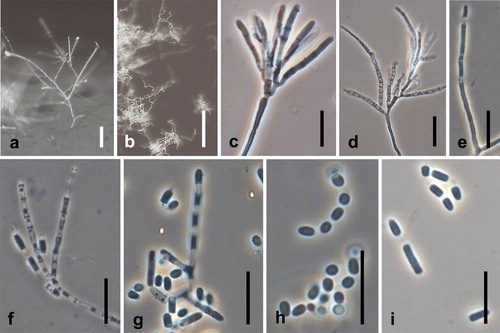
Fig. 11. Spiromastix minimus. Ex-type culture on WA. A. Submerged or aerial conidiophores. B. Conidiophores and conidia. C, D. Terminal and single lateral conidia and arthroconidia. Bars: A = 500 μm, B–D = 10 μm.
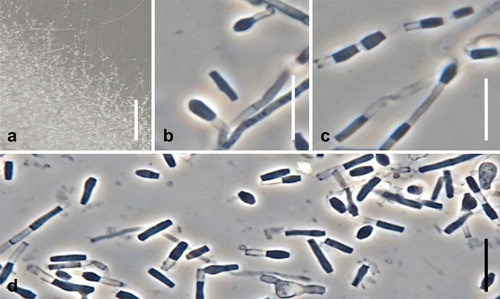
Fig. 12. Spiromastix pyramidalis. Ex-type culture on MEA. A. Aerial conidiophores. B–H. Conidiophores and conidia. I. Terminal and single lateral conidia and arthroconidia. Bars: A = 500 μm, B–G = 10 μm, H, I = 5 μm.
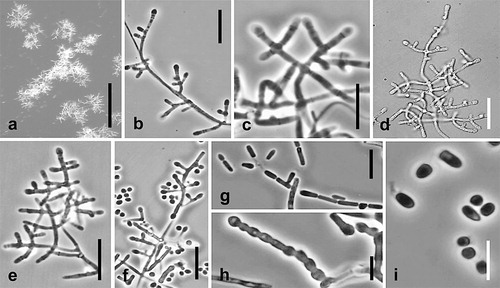
Fig. 13. Spiromastix sugiyamae. Ex-type culture on WA. A, B. Ascomata on WA. C, D. Ascomata appendages. E, F. Asci. G. Ascospores. H. Scanning electron micrograph (SEM) of ascomata. I. SEM of ascomata appendages. J. SEM of ascospores. Bars: A = 500 μm, B = 100 μm, C–E, H = 10 μm, F, G, I = 5 μm, J = 1 μm.