Figures & data
Figure 2. Cyclic diguanylic acid can bind to proteins as monomers, dimers or tetramers (dimer of a dimer).
(A) Crystal structure of cyclic diguanylic acid (c-di-GMP) bound to WspR I-site, DGC protein (protein data bank [PDB] code: 3I5A) [Citation5]; (B) Crystal structure of c-di-GMP bound to YahA, PDE protein (PDB code: 4LJ3) [Citation6]; (C) Crystal structure of c-di-GMP bound to BldD, transcriptional factor (PDB code: 4OAZ) [Citation7].
![Figure 2. Cyclic diguanylic acid can bind to proteins as monomers, dimers or tetramers (dimer of a dimer).(A) Crystal structure of cyclic diguanylic acid (c-di-GMP) bound to WspR I-site, DGC protein (protein data bank [PDB] code: 3I5A) [Citation5]; (B) Crystal structure of c-di-GMP bound to YahA, PDE protein (PDB code: 4LJ3) [Citation6]; (C) Crystal structure of c-di-GMP bound to BldD, transcriptional factor (PDB code: 4OAZ) [Citation7].](/cms/asset/5b167b79-e1a5-4867-9795-4881e2463326/ifso_a_12363885_f0003.jpg)
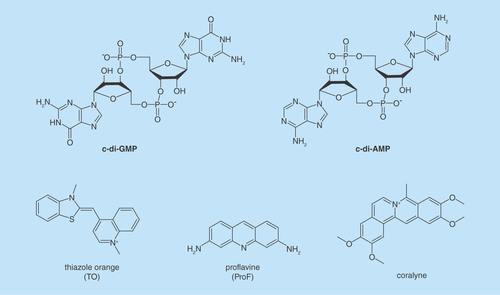
Figure 3. Different approaches to inhibit dinucleotide hydrolysis.
(A) Traditional approach to inhibiting c-di-NMP signaling; (B) unexplored approach to inhibiting c-di-NMP signaling via aggregate formation. ‘N’ in c-di-NMP, pNpN, NMP, NDP, NTP or cNMP refers to guanine or adenine base. White block indicates nucleobase. Blue and red shapes represent intercalators. Aggregated forms could be resistant to PDE cleavage.
c-di-NMP: Cyclic-di-nucleotide; cNMP: Cyclic nucleotide monophosphate; NDP: Nucleotide diphosphate; NTP: Nucleotide triphosphate; PDE: Phosphodiesterase; pNpN: Linear dinucleotide.
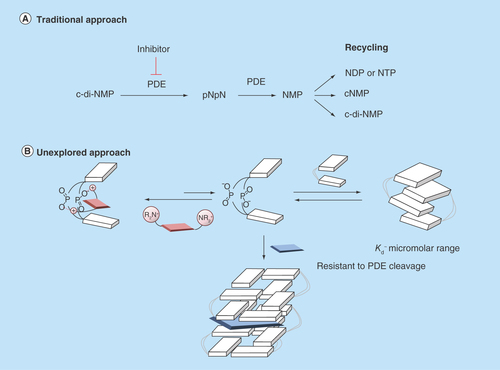
Figure 4. Proposed cyclic diguanylic acid/intercalator complexes.
(A) Tetramer/thiazole orange complex and (B) Hexamer/thiazole orange complex. Blue and white indicate guanine, M+ represents as metal ion such as K+ and colored arrows indicate fluorescence. C-di-GMP can form G-quadruplex structures and intercalators could end-stack or intercalate into these structures.
C-di-GMP: Cyclic diguanylic acid.
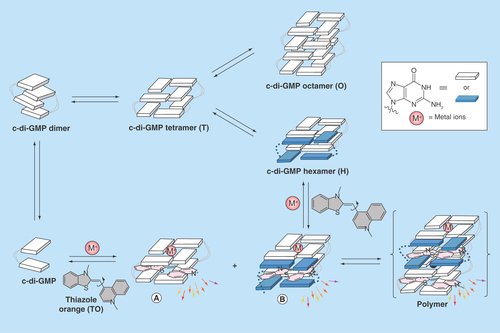
Figure 5. Complex formation between cyclic diguanylic acid and 3,6-diaminoacridine hydrochloride captured by atomic force microscopy image.
Condition: [c-di-GMP] = 100 μM, [ProF] = 100 μM, [K+] = 250 mM in 50 mM Tris-HCl (pH = 7.5) was left to stand overnight (12 h) and then diluted 20-times to give final concentrations of c-di-GMP (5 μM), ProF (5 μM) and K+ (12.5 mM), which was deposited on the mica plate for AFM imaging.
AFM: Atomic force microscopy; c-di-GMP: Cyclic diguanylic acid; ProF: 3,6-Diaminoacridine hydrochloride.
![Figure 5. Complex formation between cyclic diguanylic acid and 3,6-diaminoacridine hydrochloride captured by atomic force microscopy image.Condition: [c-di-GMP] = 100 μM, [ProF] = 100 μM, [K+] = 250 mM in 50 mM Tris-HCl (pH = 7.5) was left to stand overnight (12 h) and then diluted 20-times to give final concentrations of c-di-GMP (5 μM), ProF (5 μM) and K+ (12.5 mM), which was deposited on the mica plate for AFM imaging.AFM: Atomic force microscopy; c-di-GMP: Cyclic diguanylic acid; ProF: 3,6-Diaminoacridine hydrochloride.](/cms/asset/1c180afa-4bdc-4eb3-a461-dc33a20969fd/ifso_a_12363885_f0006.jpg)
Figure 7. Electrophoretic mobility shift assays using native gel.
100 μM unlabeled cyclic diguanylic acid (c-di-GMP) and 0.8 nM 32P-c-di-GMP were incubated with various concentrations of ProF in the presence of 250 mM K+. Final concentration of ProF: lane 1 = 0 μM; lane 2 = 1 μM; lane 3 = 5 μM; lane 4 = 10 μM; lane 5 = 15 μM; lane 6 = 20 μM; lane 7 = 30 μM; lane 8 = 40 μM; lane 9 = 50 μM and lane 10 = 100 μM. (a) = high molecular weight aggregate and (b) = monomeric c-di-GMP.
ProF: 3,6-Diaminoacridine hydrochloride.
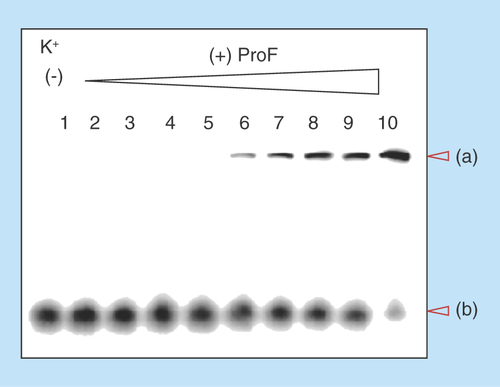
Figure 8. Fluorescence melting of the complex formed between cyclic diguanylic acid and thiazole orange or 3,6-diaminoacridine hydrochloride in the presence of different cations.
Condition: [c-di-GMP] = 100 μM, [ProF or TO] = 10 μM, [Na+, K+ or Li+] = 250 mM in 50 mM Tris-HCl (pH = 7.5). The pH of Tris is known to slightly fluctuate with temperature. As a control, melting experiment was also performed in MOPS buffer and similar melting trend was also observed, see Supplementary Figure 6. The melting of ProF itself as control was included in Supplementary Information (see Supplementary Figure 7). (A) Melting of c-di-GMP/ProF polymer in the presence of Li+; (B) melting of c-di-GMP/ProF polymer in the presence of Na+; (C) melting of c-di-GMP/ProF polymer without added cation; (D) melting of c-di-GMP/ProF polymer in the presence of K+; (E) melting of c-di-GMP/TO polymer in the presence of Na+; (F) melting of c-di-GMP/TO polymer in the presence of K+.
c-di-GMP: Cyclic diguanylic acid; ProF: 3,6-Diaminoacridine hydrochloride; TO: Thiazole orange.
![Figure 8. Fluorescence melting of the complex formed between cyclic diguanylic acid and thiazole orange or 3,6-diaminoacridine hydrochloride in the presence of different cations.Condition: [c-di-GMP] = 100 μM, [ProF or TO] = 10 μM, [Na+, K+ or Li+] = 250 mM in 50 mM Tris-HCl (pH = 7.5). The pH of Tris is known to slightly fluctuate with temperature. As a control, melting experiment was also performed in MOPS buffer and similar melting trend was also observed, see Supplementary Figure 6. The melting of ProF itself as control was included in Supplementary Information (see Supplementary Figure 7). (A) Melting of c-di-GMP/ProF polymer in the presence of Li+; (B) melting of c-di-GMP/ProF polymer in the presence of Na+; (C) melting of c-di-GMP/ProF polymer without added cation; (D) melting of c-di-GMP/ProF polymer in the presence of K+; (E) melting of c-di-GMP/TO polymer in the presence of Na+; (F) melting of c-di-GMP/TO polymer in the presence of K+.c-di-GMP: Cyclic diguanylic acid; ProF: 3,6-Diaminoacridine hydrochloride; TO: Thiazole orange.](/cms/asset/b4c9bb8d-e04c-4293-9386-e253ed789f5a/ifso_a_12363885_f0009.jpg)
Figure 9. Kinetics of cyclic diadenylic acid/coralyne complex formation.
Condition: C-di-AMP (10 μM), KI (3 mM) and buffer (50 mM Tris-phosphate [pH 7.5]) were heated up at 95°C for 5 min. Coralyne (10 μM) was then added and mixed well. The sample was subject to fluorescence monitoring at 25°C over 5 min. Excitation: 420 nm and emission: 475 nm.
C-di-AMP: Cyclic diadenylic acid.
![Figure 9. Kinetics of cyclic diadenylic acid/coralyne complex formation.Condition: C-di-AMP (10 μM), KI (3 mM) and buffer (50 mM Tris-phosphate [pH 7.5]) were heated up at 95°C for 5 min. Coralyne (10 μM) was then added and mixed well. The sample was subject to fluorescence monitoring at 25°C over 5 min. Excitation: 420 nm and emission: 475 nm.C-di-AMP: Cyclic diadenylic acid.](/cms/asset/c89d2afb-e695-4eec-89dd-d06285800785/ifso_a_12363885_f0010.jpg)
Figure 10. Intercalator-mediated dinucleotide aggregation inhibits c-di-NMP cleavage.
(A) YybT cleavage of 32P-c-di-AMP in the presence and absence of coralyne. (B) YybT cleavage of 32P-c-di-GMP in the presence and absence of ProF. Condition: [YybT] = 1.5 μM, [c-di-GMP or c-di-AMP] = 10 μM (32P labeled + unlabeled), [coralyne or ProF] = 10 or 30 μM, [KCl] = 10 mM in (B). (A) was done at 37°C for 20 min and (B) was done at 37°C for 30 min.
c-di-AMP: Cyclic diadenylic acid; c-di-GMP: Cyclic diguanylic acid; ProF: 3,6-Diaminoacridine hydrochloride.
![Figure 10. Intercalator-mediated dinucleotide aggregation inhibits c-di-NMP cleavage.(A) YybT cleavage of 32P-c-di-AMP in the presence and absence of coralyne. (B) YybT cleavage of 32P-c-di-GMP in the presence and absence of ProF. Condition: [YybT] = 1.5 μM, [c-di-GMP or c-di-AMP] = 10 μM (32P labeled + unlabeled), [coralyne or ProF] = 10 or 30 μM, [KCl] = 10 mM in (B). (A) was done at 37°C for 20 min and (B) was done at 37°C for 30 min.c-di-AMP: Cyclic diadenylic acid; c-di-GMP: Cyclic diguanylic acid; ProF: 3,6-Diaminoacridine hydrochloride.](/cms/asset/ac46868d-a9d0-457b-a40a-bd11a28ca5c4/ifso_a_12363885_f0002.jpg)