Figures & data
Figure 1. Gene ontology of 51 hypothetical proteins in Chlamydia abortus.
The proteins are classified based on biological, cellular and molecular functions.
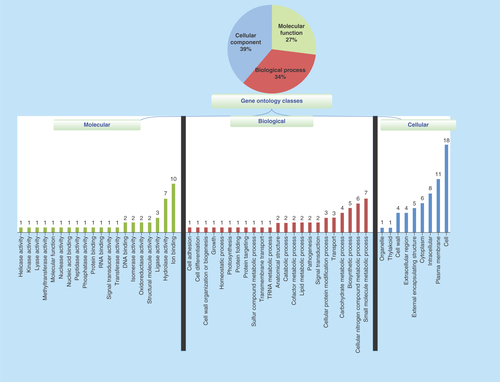
Figure 2. Structural elucidation of two proteins from Chlamydia abortus genome sequence.
(A) WP_006344020.1 (phosphorylase) and (B) WP_006344325.1 (CPAF). The amino acid residues involved in enzymatic activity are shown in red.
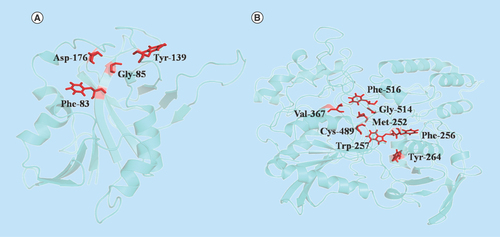
Figure 3. Molecular dynamic analysis of two homology-modeled structures showed stability in the root-mean-square deviations and radius of gyration.
Also the structural flexibility of proteins were analyzed by RMSF of the amino acid residues. Steadiness of proteins were analyzed by SASA during the MD simulations that revealed that the proteins were stable. (A) RMSD of two homology-modeled proteins were plotted, initially both the proteins showed an increase in RMSD and after 2.5 ns, proteins attained stability and consistency in structural behavior, indicating the stability of proteins during MD simulation. (B.I) WP_006344020.1 and (B.II) WP_006344325.1. The proteins had initially less compactness in structures, but at later stages, the proteins showed more compactness and less variation in the radius of gyrations along all spatial axes. (C) Protein WP_006344020.1 (blue) and WP_006344325.1 (red) with amino acid residues, 198 and 565, respectively, showed evenness in the RMSF of amino acid residues of both the proteins with the progression of MD. The uniformity indicates that the proteins are stable. (D) The SASA of the two modeled proteins was constant throughout the MD simulations.
MD: Molecular dynamics; RMSD: Root-mean-square deviations; RMSF: Root-mean-square fluctuations; SASA: Solvent surface accessible area.
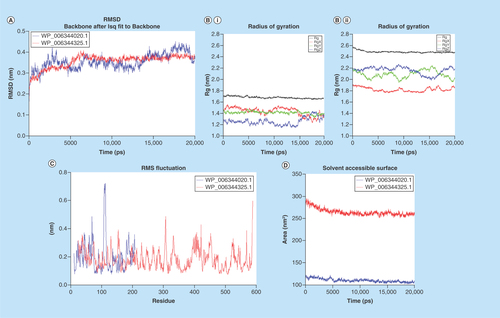
Table 1. List of bioinformatics programs used for functional analysis of hypothetical proteins in Chlamydia abortus.
Table 2. List of sequence-based analysis of 51 hypothetical proteins, gene IDs and some of their specific functions in Chlamydia abortus.
Table 3. List of hypothetical proteins of Chlamydia abortus that were showing sequence homology with selected six host proteins.
