Figures & data
Table 1. Phenotypic and genomic characteristics of nitrofurantoin resistance mechanisms of the Klebsiella pneumoniae isolates.
Table 2. Phenotypic and genomic characteristics of nitrofurantoin resistance mechanisms of the Enterobacter species isolates.
Table 3. Phenotypic and genomic characteristics of nitrofurantoin resistance mechanisms of the Citrobacter freundii, Escherichia coli and Klebsiella michiganensis isolates.
Figure 1. Evolutionary relationship of nitrofurantoin-resistant Klebsiella pneumoniae strains from different parts of the world.
Phylogenomic tree of (A)Klebsiella pneumoniae drawn with MEGA 7 and (B)K. pneumoniae and associated metadata drawn with Phandango. The isolates clustered according to clones and country of origin, although the genomic phylogeny shows a closer clustering of strains of different clones and countries. Strains of ST101 were not all of the same clade while many strains from the same hospitals and wards were of the same clade.
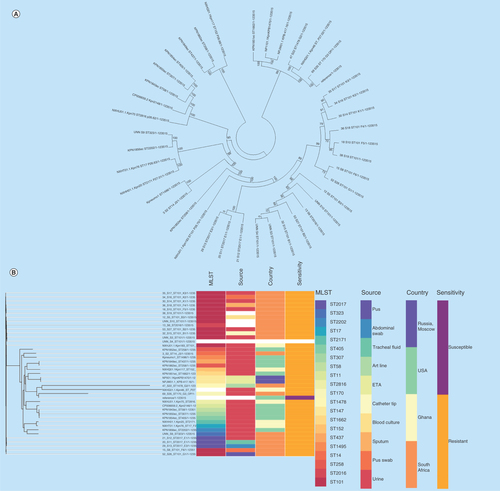
Figure 2. Phylogenomic tree of Escherichia coli nitrofurantoin-resistant strains from Belgium, USA and South Africa.
Clustering of the strains into clades were mainly country- and clone-specific.
MLST: Multi-locus sequence typing; NFT: Nitrofurantoin.
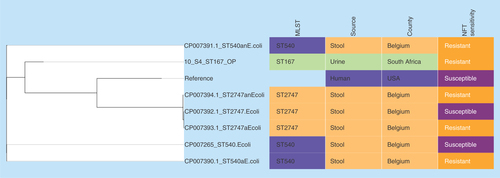
Figure 3. Phylogenomic tree of nitrofurantoin resistance Enterobacter species from South Africa, USA, Malaysia and Ghana.
Clustering of the strains into clades were mainly country- and clone-specific except for 65_S32 of ST436 from South Africa and NXHI01.1 of ST455 (Ghana), and between 1_S1 of ST108 and the Enterobacter spp. strain UNN42_S6.
MLST: Multi-locus sequence typing; NFT: Nitrofurantoin.

Figure 4. Phylogenomic tree of Citrobacter freundii nitrofurantoin intermediate resistant strains from South Africa.
Clustering of the strains into clades were mainly country- and clone-specific.
MLST: Multi-locus sequence typing; NFT: Nitrofurantoin.
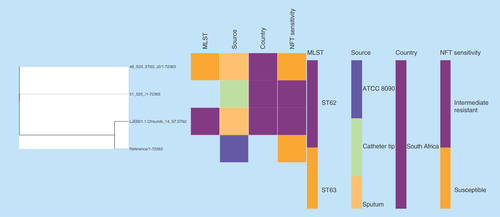
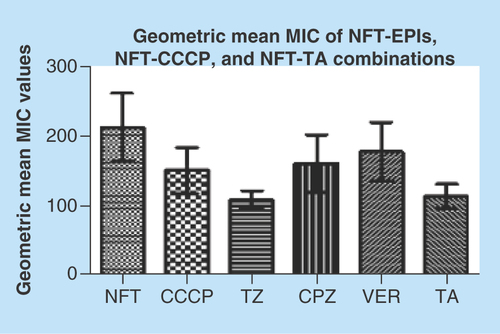
Figure 5. Geometric mean of nitrofurantoin, nitrofurantoin-carbonyl cyanide m-hydrophenylhydrazine, nitrofurantoin-tannic acid and nitrofurantoin-efflux pump inhibitor minimum inhibitory concentrations.
TZ and TA significantly reduced the geometric mean MIC of NFT (p < 0.001) than the remaining efflux-inhibiting agents.
CCCP: Carbonyl cyanide m-hydrophenylhydrazine; CPZ: Chlorpromazine; EPI: Efflux pump inhibitor; MIC: Minimum inhibitory concntration; NFT: Nitrofurantoin; TA: Tannic acid; TZ: Thioridazine; VER: Verapamil.