Figures & data
Figure 1. Expression of ZO proteins along the kidney tubule. (A) Cartoon depicting the glomerulus and various regions of the nephron and collecting duct. PCT/S1, S1 segment of the proximal tubule; TAL, thick ascending limb of Henle's loop, CCD, cortical collecting duct; OMCD, outer medullary collecting duct. (B) mRNA expression levels of ZO-1, ZO-2, ZO-3 and AQP2, used as a collecting duct-specific marker, were compared by Real-Time PCR between segments of isolated kidney tubules. Data is represented as fold difference of mRNA expression over values obtained in cortical collecting duct and is expressed as the mean ± SEM of data from 3 animals. (C) and (D) Confocal z-stacks of ZO-1, ZO-2 and ZO-3 immunostaining of the cortex, outer medulla and inner medulla of kidney slices (C) and dense mCCDcl1 cells (D). Images are representative of those acquired from 3 animals and at least 3 in vitro experiments. Bar, 10 μm. Glomeruli are indicated by an asterisk. (E) Western blot of whole-cell ZO-1, ZO-2 and ZO-3 protein depicting their expression in dense mCCDcl1 cells.
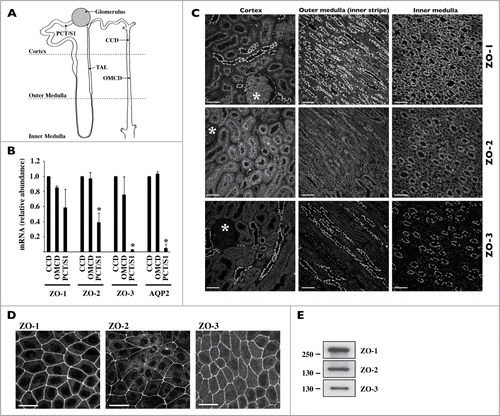
Figure 2. Characterization of mCCDcl1 cell proliferation. Cells were seeded at day 1 (D1) as described in Materials and Methods and various parameters of cell proliferation were examined over time (D1 - D9). (A) Cell number was estimated by trypsinizing and counting cells with a hemocytometer. Cell diameter was estimated by ImageJ analysis of images taken prior to cell trypsinization. Data is represented as fold increase of cell number (black squares) and cell area (red squares) over values obtained 3 d (D3, for cell number analysis) and 6 d (D6, for cell area analysis) after seeding. (B) Cell cycle analysis by flow cytometry. Data shown is representative of one of 3 similar experiments. (C) Western blot of whole-cell CycD1 and PCNA. β-actin was used as a loading control. Quantification of data, shown at right, is represented as fold difference of protein expression over values obtained at D3 and is expressed as the mean ± SEM of 3 independent experiments. (D) Confocal z-stacks of CycD1 (green, left panels) and PCNA (green, right panels) depicting their nuclear expression in low (D3) and high (D7) density cells. Enlarged single-plane (sp) images of Hoechst (blue) or immunofluorescence staining of cells outlined by a white rectangle are also shown below. One of 3 similar experiments is shown. Bar, 15 μm.
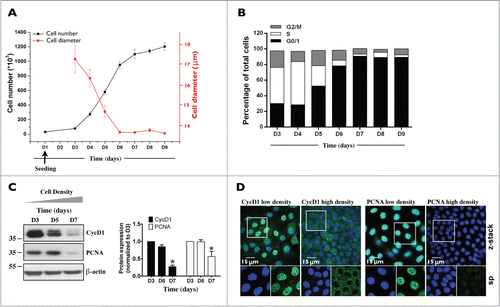
Figure 3. ZO-3 abundance and expression at junctions is dramatically increased in confluent mCCDcl1 cells. (A) Western blot of ZO-1, ZO-2, ZO-3 and ZONAB 3, 5 and 7 d (D3, D5 and D7) after seeding. β-actin was used as a loading control. Quantification of data, shown at right, is represented as fold difference of protein expression over values obtained at D5 (for ZO proteins) and D3 (for ZONAB) and is expressed as the mean ± SEM of 3 independent experiments. (B) Immunofluorescence of low- and high-density cells. Shown are confocal z-stacks of ZO-1, ZO-2, ZO-3 and ZONAB (all green) depicting their expression at junctional sites (and nuclear expression for ZONAB) in low (D3) and high (D7) density cells. Enlarged single-plane images of Hoechst (blue) or immunofluorescence staining of cells are also shown below. Bar, 10 μm. (C and D) Immunofluorescence of high-density cells 6 h after scratch. Shown are confocal z-stacks of PCNA (green) and CycD1 (green) (C), ZONAB (green) (D) and double-stained confocal z-stacks of ZO-1 (red) / ZO-2 (green) and ZO-1 (red) / ZO-3 (green) (D), depicting their expression at junctional sites (and nuclear expression for PCNA, CycD1 and ZONAB) of cells bordering scratch wounds and of cells located at more distant regions. Enlarged images of Hoechst (blue) or immunofluorescence staining of cells bordering scratch wounds are also shown below. Bar, 30 μm. For (B–D), one of 3 similar experiments is shown.
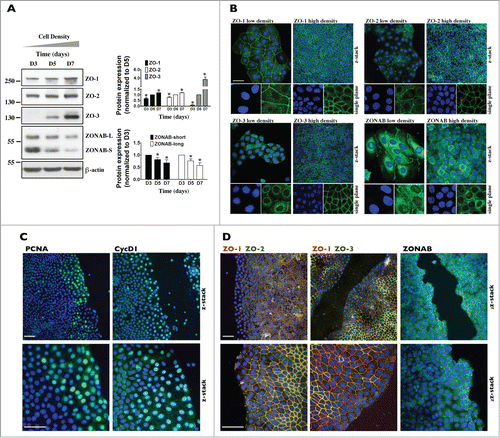
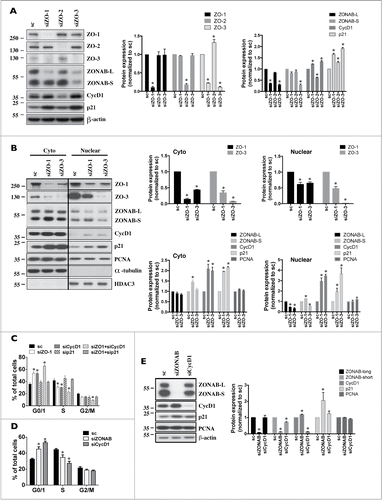
Figure 4. ZO-3 protein expression relies on the presence of ZO-1 in mCCDcl1 cells. (A and B) Cells were transfected 1 day after seeding with siRNA (sc) or siRNA against ZO-1 (siZO-1), ZO-2 (siZO-2) or ZO-3 (siZO-3) and mRNA (A) and protein (B) were isolated 1 - 6 d post-transfection (T1 - T6). Data is represented as fold difference of mRNA expression over values obtained in cells transfected with scrambled siRNA at T1 (for mRNA analysis) or at T2 (for protein analysis) and is expressed as the mean ± SEM of 4 independent experiments. Significant change of expression (P ≤ 0.05) within a same experimental group is depicted by an asterisk. (C) Confocal z-stacks of ZO-1, ZO-2 and ZO-3 depicting their expression in cells transfected with either scrambled siRNA, siZO-1, siZO-2 or siZO-3 at T3. One of 3 similar experiments is shown. Bar, 10 μm.
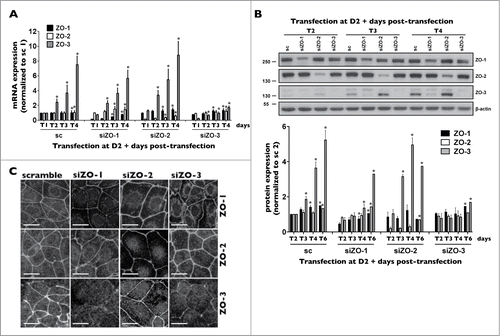
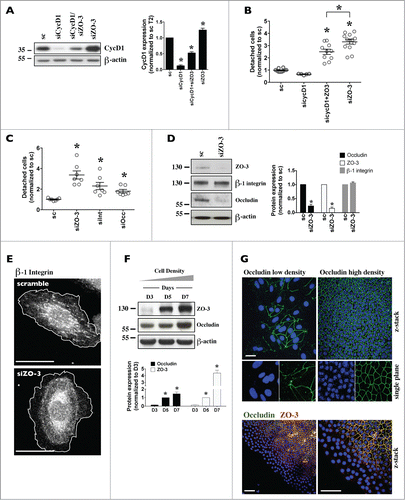
Figure 5. Decreased expression of each ZO protein differently affects mCCDcl1 cell cycle progression and adhesion. (A-C) Cells were transfected 1 day after seeding with scrambled siRNA (sc) or siRNA against ZO-1 (siZO-1), ZO-2 (siZO-2) or ZO-3 (siZO-3) and cell number (A), cell confluence (B) and cell volume (C) were measured 1–4 d post-transfection (T1 - T4). Data is expressed as the mean ± SEM of 4 independent experiments. Cell number is represented as fold difference of siZO data over values obtained in cells transfected with scrambled siRNA at T1. (D) Cell death analysis by flow cytometry of cells transfected with scrambled siRNA or siRNA against ZOs at T3. Staurosporine (STP, 500 nM for 24 h) was used as a positive control for cell apoptosis. (E) Cell cycle analysis by flow cytometry of cells transfected with scrambled siRNA or siRNA against ZOs at T2 (left panel) or of cells exposed to siRNA for 1 day (T1), re-seeded and analyzed 1 day later (right panel). Data is expressed as the mean percentage of G0/G1, S and G2/M phase cells ± SEM of 4 independent experiments. (F) Quantification of cell detachment by flow cytometry of cells transfected with scrambled siRNA or siRNA against ZOs. Data is represented as fold difference of detached cells over values obtained in cells transfected with scrambled siRNA at T2 and is expressed as the mean ± SEM of 5 independent experiments performed in duplicate. NS: no significant difference. (G) Phase-contrast microscopy of high-density cells (T6) illustrating the absence of domes and increased number of detached cells in cells transfected with siZO-1 and siZO-3 as compared to cells transfected with scrambled siRNA or siZO-2. Enlarged images of cells outlined by a white rectangle are shown in inserts. One of 4 similar experiments is shown. Bar, 50 μm.
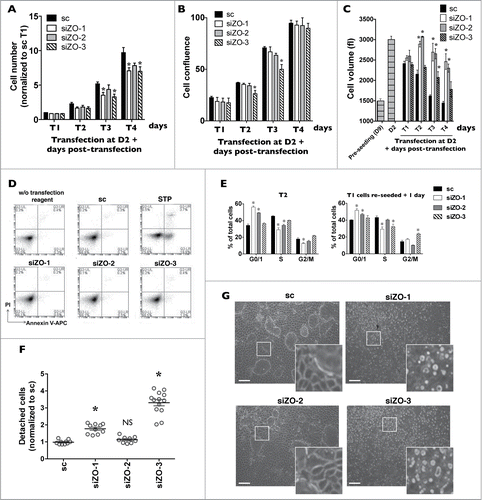
Table 1. RT-PCR primer sequences and siRNA sequences