Figures & data
Figure 1. Wild-type p53 is stabilized by proteasome inhibitors. (A and B) U2OS osteosarcoma cells and MCF7 breast cancer cells were treated with the indicated concentrations of proteasome inhibitors MG132, bortezomib (Bor), carfilzomib (Car) or thiostrepton (Thio). Immunoblotting was performed for HDM2, p53, p21. β-actin was used as the loading control.
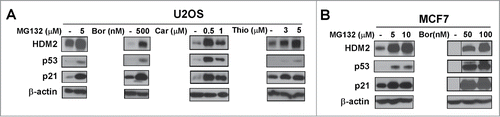
Figure 2. Mutant p53 is suppressed by proteasome inhibitors. (A–C) MDA-MB-231 breast, MIA PaCa-2 pancreatic and DU145 prostate cancer cells were treated with the indicated concentrations of proteasome inhibitors MG132, bortezomib (Bor), carfilzomib (Car) or thiostrepton (Thio). Immunoblot analysis of HDM2, p53, p21 and β-actin as the loading control was carried out after treatment.
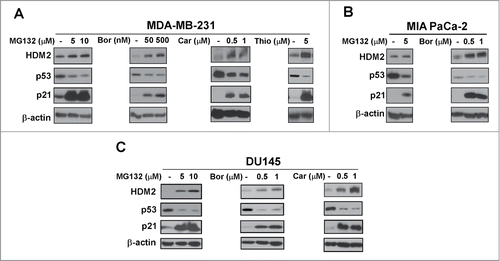
Figure 3. Arsenic trioxide acts as a proteasome inhibitor. (A) C3-luc cells were treated as indicated for overnight and luciferase activity was measured using the Luciferase Assay System kit from Promega. Graph shows mean ± SEM of 2 independent experiments. (B–D) MIA PaCa-2, DU145 and U2OS cells were treated as indicated. Immunoblotting was performed with antibodies specific for FOXM1, HDM2, p53, Mcl-1, p21 and ubiquitin. β-actin was used as the loading control.
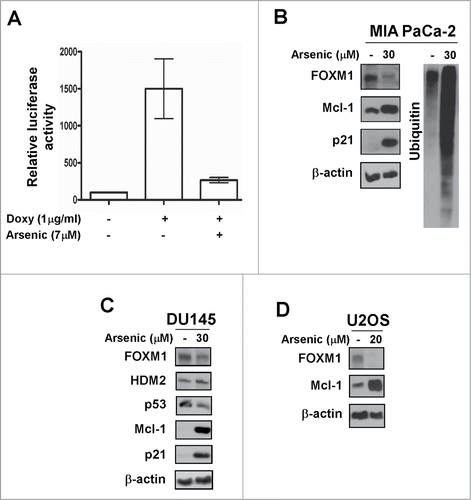
Figure 4. Proteasome inhibitor-associated suppression of mutant p53 is partially rescued by HDM2 knockdown. (A–C) MIA PaCa-2 and MDA-MB-231 cells were transfected with either control or HDM2 specific siRNA. Following a 72-hour transfection cells were treated as indicated. Cell lysates were immunoblotted for HDM2, p53, p21 and β-actin as the loading control.
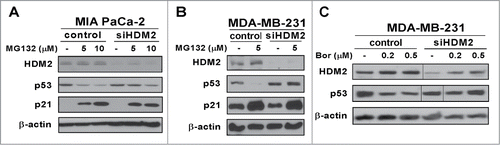
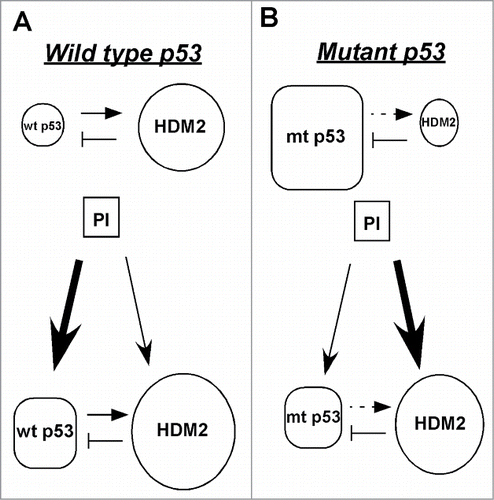
Figure 5. Model of the HDM2-mediated suppression of mutant p53 after treatment with proteasome inhibitors. (A) The basal level of wild-type p53 is low because HDM2, its transcriptional target and negative regulator marks it for proteasomal degradation. After treatment with proteasome inhibitors both wild-type p53 and HDM2 are stabilized. Though HDM2 continues to degrade wild-type p53, but its overall level increases because its degradation by HDM2 is overridden by its stabilization by proteasome inhibitors. (B) The basal level of mutant p53 is high because it cannot transactivate its negative regulator HDM2. Following proteasome inhibitor treatment both mutant p53 and HDM2 are stabilized, but the overall level of mutant p53 decreases because the increased amount of HDM2 efficiently targets it for degradation, thus its stabilization by proteasome inhibitors is overridden by its HDM2-mediated degradation.