Figures & data
Figure 1. Deletion of RFTS enhances the oncogenic activity of DNMT1. (A) HBEC3 stable cell lines were established to express full-length and DNMT1 deletion forms near endogenous DNMT1 levels. The levels of DNMT1 were determined by RT-qPCR (left) and western blotting (right). Data were normalized to vector cells. (B) Adherent colony formation. (C) Soft-agar colony formation. *, p < 0.05; **, p < 0.01; ***, p < 0.001; ns, indicates no significant difference in comparison to vector cells.
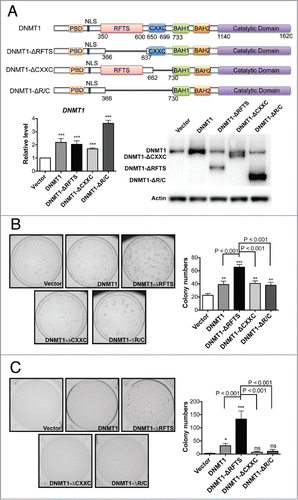
Figure 2. DNMT1-ΔRFTS promotes increased methylation and silencing of the DAPK and DUOX1 genes. (A) The methylation levels of DAPK (left) and DUOX1 (right) promoter-associated CpG islands were analyzed by qPCR. Methylated DNA was analyzed using the MethylMiner kit and amplified with specific primers. (B) Bisulfite sequencing results for DAPK (left) and DUOX1 (right) promoters. White squares represent unmethylated cytosines and black squares represent methylated cytosines in CpG sites. The percentage of methylated CpG dinucleotides from 8 independent clones is indicated. (C) DNMT1 chromatin occupancy was analyzed using DNMT1 ChIP and qPCR. (D) mRNA levels of DAPK (left) and DUOX1 (right) were analyzed by RT-qPCR and normalized to vector cells. *, p < 0.05; **, p < 0.01; ***, p < 0.001 in comparison to vector cells.
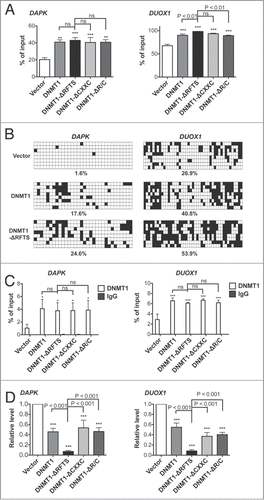
Figure 3. DNMT1-ΔRFTS decreases chromatin accessibility at DAPK and DUOX1 promoters. Cells were treated with or without DNA nuclease for 1 hr, prior to detection of promoter DNA by qPCR. The index of chromatin accessibility = 2 ((Ct DNase treated)-(Ct Untreated)). *, p < 0.05; ***, p < 0.001 in comparison to vector cells.
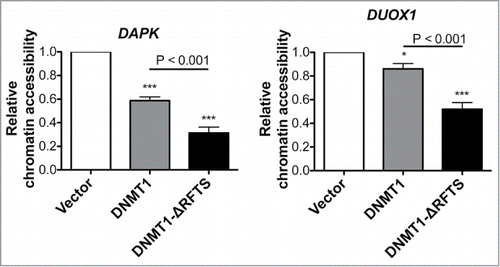
Figure 4. 5-aza-dC treatment reactivates TSG expression and suppresses DNMT1-dependent transformation. (A) mRNA levels of DAPK (left) and DUOX1 (right) were analyzed by RT-qPCR after 100nM 5-aza-dC treatment for 5 d and normalized to vector cells treated with DMSO. (B) Soft-agar colony formation after 5-aza-dC treatment. ***, p < 0.001 in comparison to the DMSO treated control.
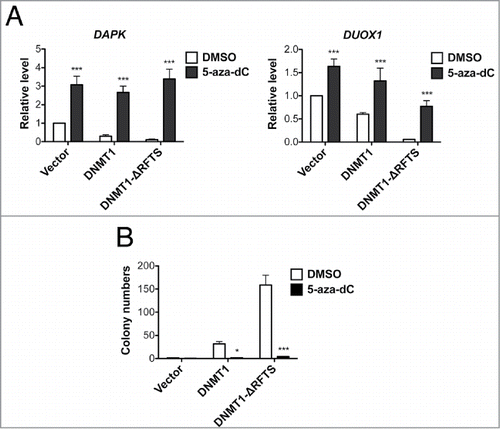
Figure 5. DNMT1-ΔRFTS expression enhances global DNMT1 methylation changes. (A) Genome-wide promoter DNA methylation profiles were obtained using the HELP assay. In volcano plots, the x-axis scores probe-specific methylation ratios and the y-axis scores p-values for the confidence of measurements. The plots allow visualization of methylation differences between vector and DNMT1 cells as well as the differences between vector and DNMT1-ΔRFTS cells. Probes sets that showed significant hyper- or hypomethylation (p < 0.05 for methylation changes (log2(HpaII/MspI)) > 2) are shown in cyan. All other probes are shown in red. (B) Heat map illustration of HpaII-enrichment fragments with methylation changes (log2(HapII/MspI)) > 2 between vector and DNMT1-ΔRFTS cells.
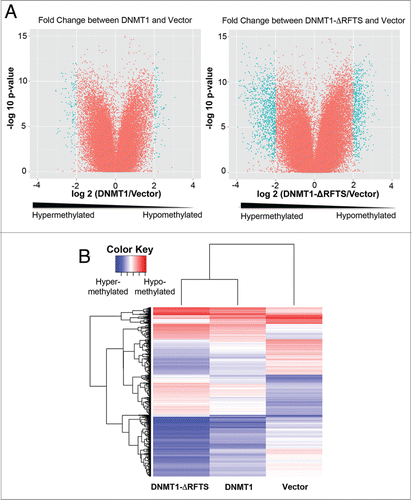
Figure 6. Genomic hypomethylation is found in DNMT1-ΔRFTS cells. (A) 5-methylcytosine (mC) content of the total cytosine pool was determined by HPLC. (B) Bisulfite sequencing of SAT2. White squares represent unmethylated CpGs, black squares represent methylated CpGs, and gray squares represent undetermined sites. Each row is an independent sequencing result. (C) Quantitation of SAT2 bisulfite sequencing. (D) DNMT1 chromatin occupancy was analyzed using DNMT1 ChIP and qPCR. (E) Expression of SAT2 non-coding RNA was analyzed by RT-qPCR and normalized to vector cells. *, p < 0.05; **, p < 0.01; ***, p < 0.001 in comparison to vector cells.
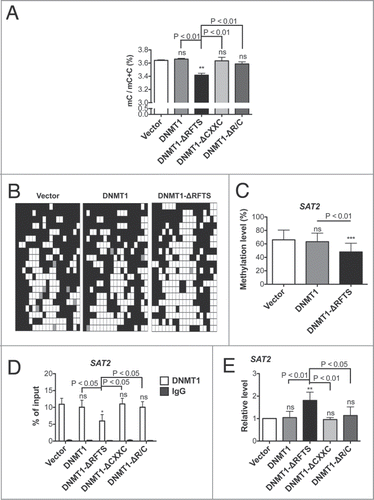
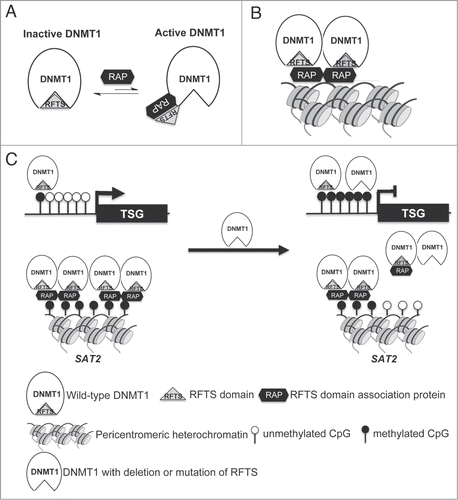
Figure 7. Dual roles for RFTS domain in DNMT1-dependent DNA methylation. (A) RFTS-targeted DNMT1 associated proteins (RAP) are proposed to relieve inhibition of DNMT1 for access to euchromatin. (B) The RFTS domain mediates association between DNMT1 and pericentromeric heterochromatin. (C) In cancer, overexpression of RAPs or mutation of RFTS is proposed to relieve DNMT1 inhibition, thereby increasing methylation and silencing of TSGs. However, because the RFTS domain is required for association with heterochromatic SAT2 sequences, DNMT1 with mutant RFTS may be less associated with such sequences, accounting for global hypomethylation.