Figures & data
Figure 1. MiR-214 increases DNA damage-induced chromosomal instability. Cells were treated with 5 Gy IR 66 h after transfection and micronuclei were detected 20 h later. (A) Four types of SKOV3 cells were observed on the basis of micronuclei: without micronucleus, with centromeric micronucleus, with non-centromeric micronucleus and with both types of micronuclei. Green, Human centromeres; Red, DNA; Bars = 10 μm. Non-centromeric (white arrows) or centromeric (yellow arrows) micronucleus. (B and C) The percentage of cells with non-centromeric micronuclei in SKOV3 (B) or OV2008 (C) cells. n = the number of cells counted. Mean ± SD, from two independent experiments. *P < 0.05, chi-square test.
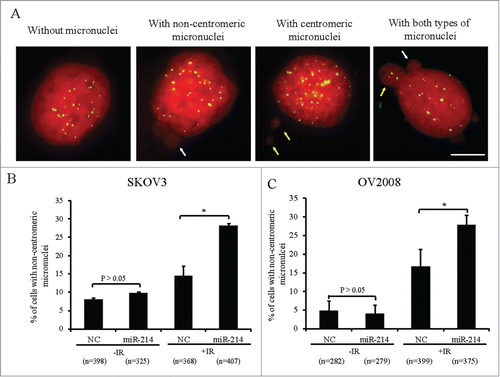
Figure 2. MiR-214 directly regulates RNF8 expression. Relative expression levels of miR-214 (A), RNF8 mRNAs (B) or protein (C and D) were detected at 48 h or 72 h post transfection by real-time PCR or Western blotting, respectively. (E) Mature miR-214 sequence and its putative binding sites in the 3′ UTR of human RNF8 mRNA. Dual luciferase reporter constructs contain a DNA sequence encoding wild-type (E1) or mutant RNF8 mRNA 3′ UTR for each individual (E2-4) or all (E5) putative binding sites, respectively. Mean ± SD, from 3 independent experiments. (F) Relative luciferase activity was measured 24 h after co-transfection of each luciferase vectors (100 ng) together with miR-214 mimics (50 nM) or controls in 293T cells. *P < 0.05, ***P < 0.001, 2-tailed t-test (B and D), chi-square test (F).
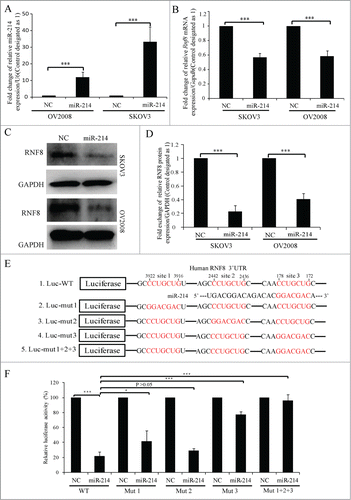
Figure 3. MiR-214 disturbs DNA damage response by downregulating RNF8 expression. A hundred nM MiR-214 mimics or RNF8 siRNAs were transfected individually or together into SKOV3 cells. Forty 8 or 72 h later, relative RNF8 mRNA (A) or protein (B) expression was detected by real-time PCR or Western blotting. The expression of downstream proteins 53BP1, BRCA1 or Rad51 in DNA damage response was also detected by Western blotting (C). Cells were treated with 5 Gy IR 72 h after transfection and then fixed for immunofluorescent staining for RAD51 and γH2AX. Representative images (D) and quantitative results (E) of RAD51 and γH2AX staining. Green,γH2AX; RAD51, Red; Blue, DNA; Bars = 10 μm. One-2 hours following treatment of cells with 5 Gy IR 72 h after transfection, neutral comet assay was performed to detect DNA damage. (F) Quantitative results were calculated by olive moment. n = the number of cells counted. ***P < 0.001, chi-square test (D), two-tailed t-test (A and F).
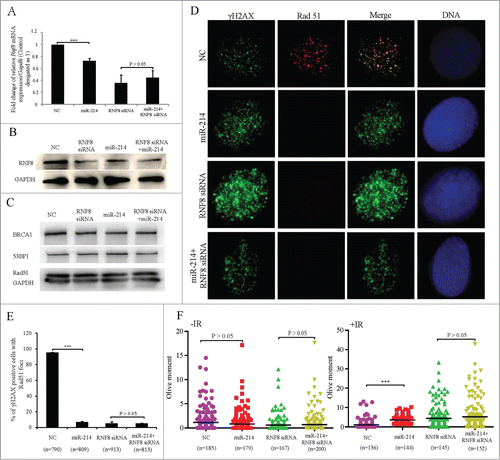
Figure 4. Inhibition of endogenous miR-214 increases RNF8-mediated DNA damage response. Representative image (A) and quantitation (B) of RNF8 expression at 72 h after miR-214 inhibitor transfection detected by Western blotting. (C) At 72 h after transfection, neutral comet assay was performed to detect DNA damage and representative images were shown, Bars = 20 μm. (D) Quantitative results were calculated by olive moment. n = the number of cells counted. Seventy-2 hours post transfection, the percentage of cells with >10 γH2AX foci were analyzed at different time points after IR treatment. (E) Representative images of γH2AX staining, Bars = 10 μm and (F) quantitative results of γH2AX staining. More than 100 cells were analyzed for each category. Seventy-2 hours post transfection, 1 × 103 A2780 cells transfected with miR-214 inhibitors or controls were treated with 5 Gy IR and followed by clone formation assay, respectively. Representative image (G) and quantitative analysis (H) of clone formation assay 7 days after IR. A2780 cells were transfected with or without miR-214 inhibitors for 72 h and followed by 5 Gy IR treatment. *P < 0.05, ***P < 0.001, chi-square test (F and H), two-tailed t-test (B and D). Mean ± SD, from two independent experiments.
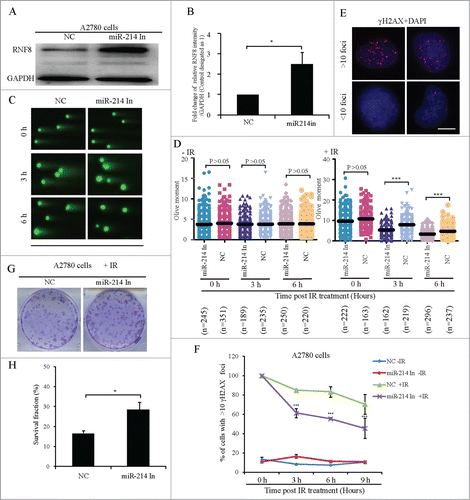
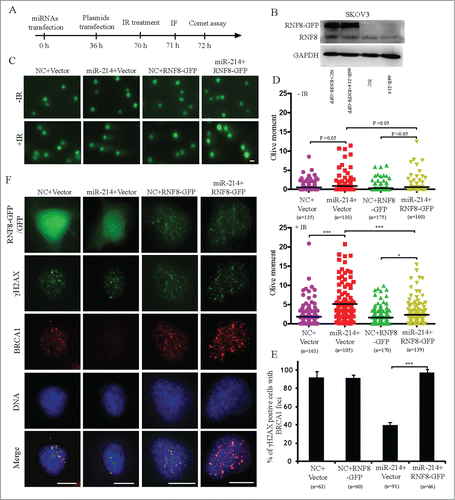
Figure 5. Overexpression of RNF8 mRNAs without 3′ UTR can antagonize the effects of miR-214 on DNA damage response. (A) Experimental scheme for cell transfection. Thirty-6 hours after transfection either with negative control or miR-214 mimics, SKOV3 cells were transfected with plasmids with or without RNF8-GFP cDNA lacking the 3′ UTR. Western blotting was performed 36 h later after plasmids transfection to detect RNF8 expression (B). Cells were then treated with 5 Gy IR and followed by recovery for about 1 or 2 h followed by immunofluorescent staining or neutral comet assay to detect DNA damage. Representative images (C) and Quantitative analysis (D) of comet assay. n = the number of cells counted. Bars = 20 μm. (E) Representative images and (F) quantitative results of RAD51 and γH2AX staining in various experimental treatments. Bars = 10 μm, Mean ± SD, from two independent experiments. *P < 0.05, ***P < 0.001, 2-tailed t-test (D) and chi-square test (F).