Figures & data
Figure 1. Relationship between expression of p12, incorporation of EdU and cellular DNA content. A549 cells were exposed to EdU for 30 (top rows) or 60 min (bottom rows), the EdU incorporation was detected by the Click-ItTM protocol, cellular expression of p12 was detected immunocytochemically, DNA was counterstained with DAPI and cellular fluorescence was measured by laser scanning cytometry.Citation60-62 During the “paint-a-gate” analysis the cells incorporating EdU were colored red (A and E) and on the bivariate distribution histograms the EdU incorporation is correlated with expression of p12 presented either as the mean integrated value of fluorescence intensity over cell nucleus (B and F) or the mean intensity of maximal pixel (C and G). The dashed skewed lines in (B, C, F, and G) show the upper level of fluorescence of the cells stained with the secondary Ab only; the cells below these lines are thus considered p12 negative. Also presented is a direct relationship between p12 and EdU incorporation (D and H). The inset in (D) shows the DNA content frequency histogram from that culture. The dashed lines in (E) outline the cells that during duration of the 60 min pulse-exposure to EdU were entering S (eS), were constantly present during pulse duration (mid-S), or were entering G2 (eG2). It is quite evident that nearly all cells with DNA content equivalent of S are incorporating EdU and are p12 negative. Presented is the percentage of cells at the G1 to S transition (with DNA content equivalent of G1) that are p12-negative and did not incorporate EdU (42% and 33%; pointed at by open arrows). As described in the text, such distribution of EdU-negative and p12-negative cells at the G1 to S transition is indicative that initiation of DNA replication starts rather rapidly after loss of p12; the amount of DNA synthesized at that time however is so small that based on DNA content these cells are still recognized as in G1. Likewise among the p12 negative cells at the S/G2 transition (with a G2M DNA content) also less frequent are the cells that did not incorporate EdU (34% and 29%). Thus termination of DNA replication appears to be shortly followed by re-expression of p12.
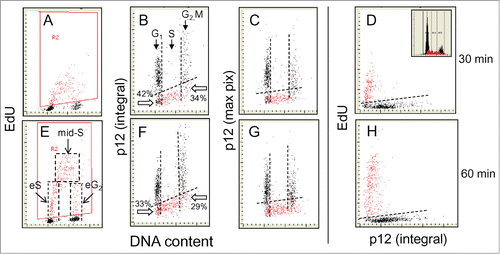
Figure 2. Relationship between expression of p21WAF1 and EdU incorporation. Similar as in the cells were exposed to EdU for 30 or 60 min, the EdU labeled cells were gated (colored red; A and E) and the EdU incorporation is correlated with expression of p21 and cellular DNA content (B, C, F, and G). Also presented is a direct relationship between p21 and EdU incorporation (D and E). The inset in (D) shows DNA content histogram of cells from this culture. The details on gating strategy and “paint-a-gate” data analysis are described in the legend to ; the dashed skewed lines represent the top level of fluorescence of the cells stained with the secondary Ab only and thus discriminate between the p21 negative and positive cells. As is evident, essentially all cells incorporating EdU are p21 negative. Among the p21-negative cells at the G1 to S transition during the 30 and 60 min EdU pulse there are 76% and 80% cells that did not incorporate EdU. Likewise at the S to G2 transition majority of cells (63% and 57%) were not incorporating EdU.
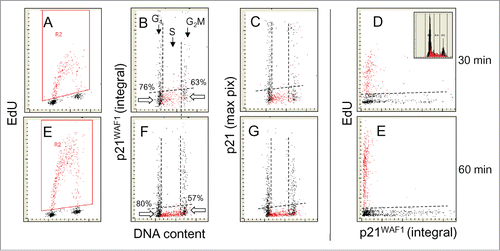
Figure 3. Relationship between expression of Cdt1 and EdU incorporation. Similar as in , the cells were exposed to EdU for 30 or 60 min, the EdU labeled cells were gated (colored red; A and E) and the EdU incorporation is correlated with expression of Cdt1 and cellular DNA content (B, C, F, and G). Also presented is a direct relationship between Cdt1 and EdU incorporation (D and E). The inset in (D) shows DNA content histogram of cells from this culture. The details on gating strategy and “paint-a-gate” data analysis are described in the legend to ; the dashed skewed lines represent the top level of fluorescence of the cells stained with the secondary Ab only and thus discriminate between the Cdt1 negative and positive cells. As is evident, essentially all cells incorporating EdU are Cdt1 negative. Among the Cdt1-negative cells at the G1 to S transition the cells that did not incorporate EdU are 55% and 46% for the 30 and 60 min EdU pulses and 56% and 55% at the S to G2 transition.
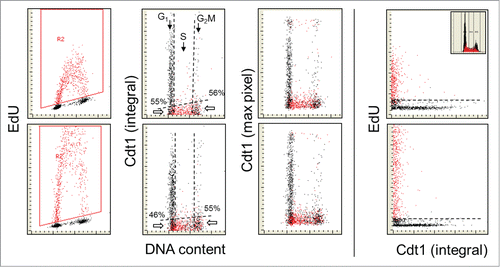
Figure 4. Relationship between expression of cyclin A and EdU incorporation. The cells were exposed in culture to EdU for 60 min. As in the EdU incorporating cells are gated/colored red (A). On the bivariate distributions of cyclin A versus DNA content (B and C) it is evident that nearly all cells incorporating EdU are cyclin A positive and that cell advancement through the S phase is associated with the progressively rising expression of this protein. In accordance with our prior findingsCitation41,42,64 cells in mitosis (M) are cyclin A negative (they were identified by the image analysis on the iCys cytometer)Citation62,63 while G2 cells have maximal level of this protein (B). As described in the text the correlation between the extent of EdU incorporation and expression of cyclin A among the mid-S phase cells, selected/gated as shown by the dashed vertical lines (A), is weak (r = 0.26; E) but stronger when all EdU-labeled cells are analyzed (r = 0.47). The DNA content histogram of these cells from this culture is shown as the inset in D.
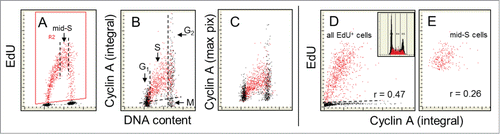
Figure 5. Relationship between expression of PCNA and EdU incorporation. The cells were exposed to EdU for 60 min and similar as in the prior figures () the EdU-labeled cells were gated, colored red (A) and analyzed with respect to their expression of PCNA as measured by the integral (B) or maximal pixel (C) intensity of nuclear fluorescence. The PCNA-negative cells are below the dashed skewed line. Note striking similarity between the patterns of EdU incorporation (A) and PCNA expression (B) in relation to DNA content. All cells that incorporate EdU are expressing PCNA. The cells just initiating DNA replication have minimal expression of PCNA. Among the mid-S phase cells that all were exposed to EdU for full 60 min (E) the correlation between expression of PCNA and the amount of incorporated EdU is relatively strong (r = 0.72) and similar (r = 0.73) when assessed for all EdU-incorporating cells. The DNA frequency histogram of these cells is shown in (D).
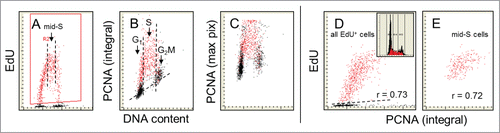
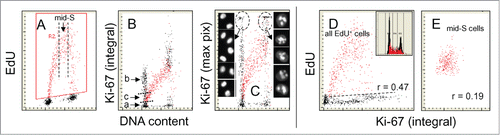
Figure 6. Relationship between expression of Ki-67 protein and EdU incorporation. The cells were exposed to EdU for 60 min, and as in the prior figures the cells incorporating the precursor are gated/colored red (A). Note marked a difference when Ki-67 fluorescence intensity is plotted as integral (B) compared to that when shown as maximal pixel (C). The high intercellular variability and intensity of fluorescence of the Ki-67 maximal pixel plot (C) is due to the punctuate localization of its strong fluorescence in nucleoli. The cells in G1 and G2 with high intensity of maximal pixel of Ki-67 were gated (marked by oval dashed lines) and their representative images are shown on both sides of (C). Note the presence of 2 distinct G1 cell subpopulations, Ki-67 negative (a) and positive (b+c). The cells initiating DNA replication have the intermediate level of Ki-67 expression (c). As in the mid-S phase cells incorporating EdU cells were gated out (vertical dashed lines in A) and their EdU incorporation was plotted vs. Ki-67 expression to assess the degree of correlation (r = 0.19) between these variables (E). This correlation was stronger when all DNA replicating cells were analyzed (r = 0.47). The DNA frequency histogram of the cells is shown in (D).