Figures & data
Figure 1. Phosphorylated Rb is localized to the mitochondria. Asynchronously growing MCF7 breast cancer cells were fractionated using a Mitochondria Isolation kit (Thermo Scientific) according to the manufacturers directions. Equal amounts of protein (15 μg) taken from the cytoplasmic/nuclear fraction and mitochondrial fractions were subjected to immunoblotting analysis as described in the Materials and Methods section. Immunodetection of Actin was used as a marker of the cytoplasmic/nuclear fraction, and detection of AIF and Bak are shown as mitochondrial localized proteins. Antibodies to total Rb and Rb phosphorylated at specific phosphorylation sites are listed to the right of the figure. Data shown is representative of two independent experiments.
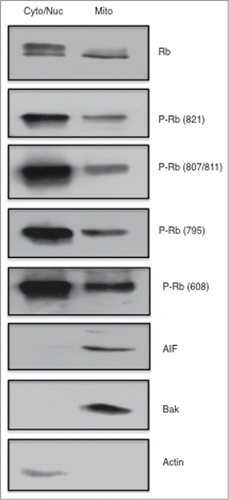
Figure 2. Rb phosphorylated on S807/S811 binds to GST-Bax. GST fusion proteins (Sigma) were used in pull down/binding assays performed as previously described (38). (A) Pull down binding assays were performed using 4 bcl2 family proteins individually fused to GST (GST-Bad, GST-Bax, GST-bcl2, and GST-Bid). Two micrograms of fusion protein was incubated with MCF7 cell lysates (500 ug) followed by analysis of GST-fusion protein associated proteins by immunoblotting with antibodies to Rb. (B) Lysates from 3 cancer cell types (MCF7, MDA-MB-231, Hs578T) were utilized in GST-Bax fusion protein binding assays followed by immunoblotting with antibodies to Rb phosphorylated at S807/S811. Data shown is representative of three independent experiments.
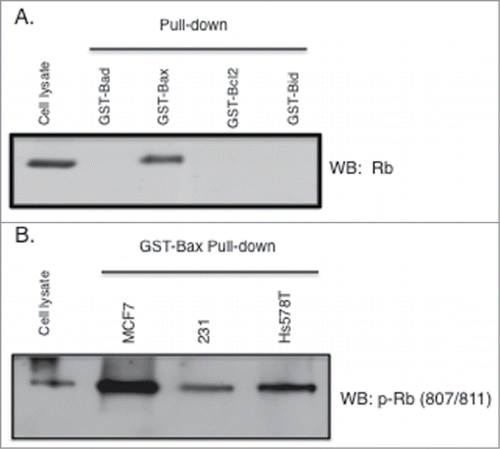
Figure 3. Association of Bax with Rb phosphorylated on S807/S811. (A) MCF7 cell lysates (1 mg) were used in immunoprecipitations with antibodies to Rb phosphorylated on S795 or S807/S811. Immunoprecipitates were analyzed by immunoblotting with antibodies to Rb or Bax. (B) MCF7 and Hs578T cell lysates were used in Bax or Rb immunoprecipitation experiments, followed by immunoblotting analysis with antibodies to phosphorylated Rb (S807/811) and Bax. Data shown is representative of three independent experiments.
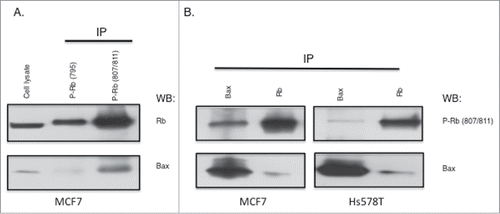
Figure 4. Rb phosphorylation at S807 is required for association with Bax. Rb mutant plasmids were generated as described in the Materials and Methods section. Rb-negative C33A cells were transfected using Fugene (Promega). (A) Rb plasmids expressing either S807A or S811A alanine mutants were transfected into cells and 48 hours later cell lysates were utilized in GST-Bax fusion protein pull down assays. Proteins associated with GST-Bax were analyzed by immunoblotting with Rb antibodies. (B) Rb plasmids expressing either S807A or S807E mutants were transfected into C33A cells and 48 hours later co-immunoprecipitation with Bax antibodies was performed. Immunoprecipitates were analyzed by immunoblotting with Rb and Bax antibodies. Equivalent expression of the Rb mutant proteins in the C33A cells is verified by immunoblotting (lower panel). Data shown is representative of three independent experiments.
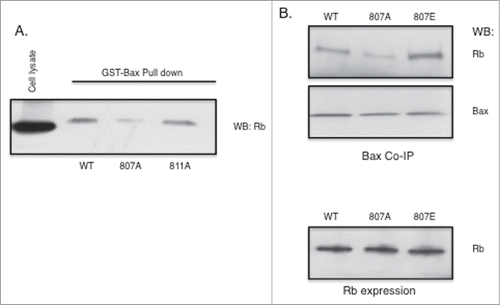
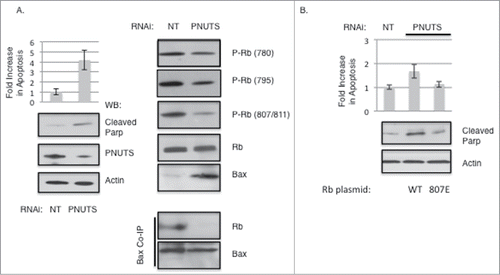
Figure 5. Dephosphorylation of Rb on S807/S811 causes Bax dissociation and stimulates Apoptosis. Knockdown of PNUTS (Phosphatase Nuclear Targeting Subunit) was performed in MCF7 cells as described in the Materials and Methods section. (A) Cells were subjected to PNUTS knockdown (PNUTS) or transfected with nontargeting RNA (NT) and 48 hours later apoptosis was measured using the Cell Death Detection ELISA (Roche Diagnostics) which detects degraded DNA released from the nucleus into the cytoplasm. The amount of apoptosis (degraded DNA) detected in control cells (NT) was normalized to one. Graph depicts the fold increase in degraded DNA observed due to PNUTS knockdown. Error bars represent standard deviation of the mean of triplicate samples and data shown is representative of three independent experiments. Apoptosis was also measured by immunoblot analysis of the cleavage of Parp as an indicator of apoptosis. Immunoblotting of Actin and PNUTS verify PNUTS knockdown. Co-immunoprecipitation experiment results and immunoblotting with site-specific Rb phosphorylation antibodies are shown on the right panel. Data shown is representative of three independent experiments. (B) C33A cells were transfected with WT or S807E Rb mutant expressing plasmids and 48 hours later PNUTS knockdown was performed. Apoptosis was measured and quantified as described in (A). Error bars represent standard deviation of the mean of triplicate samples and data shown is representative of three independent experiments.