Figures & data
Figure 1. P53 and E2F binding sites on the NANOG promoter. A 6001 bp sequence of the human NANOG gene, from nt 7936295 to nt 7942295 of the NCBI GI 224589803 (http://www.ncbi.nlm.nih.gov/nuccore/224589803), was considered the 5’-flanking Transcription Start Sequence (TSS), which encompass the −5700 + 300 gene region. JASPAR, PATCH and ALIBABA2 programs were used to identify the putative transcription-factor binding site. Panel A shows the nucleotide positions of the P53 putative and validated binding sites. Panel B shows the E2F binding sites.
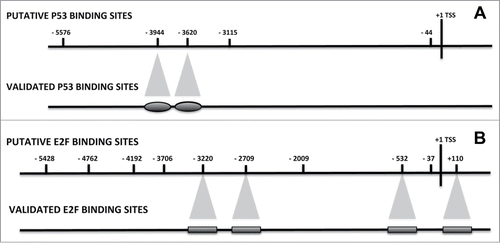
Figure 2. CHIP analysis of the P53, RB1 and RB2 binding on the NANOG promoter. MSC with silenced or over-expressed BRG1 was used for CHIP analysis. The upper graph shows CHIP results in cells with silenced BRG1. In these cells, the P53, RB1 and RB2 binding on the corresponding P53 and E2F sites, respectively, was compared with control MSC cultures. Normalized data with input percent method in cells with silenced BRG1 were divided by normalized data in control cells. Ratio values are expressed as changes in the binding level (fold change). The numbers on the X-axis refer to the nucleotide positions of the identified binding sites. The lower graph shows the same analysis in cells with overexpressed BRG1. Three biological replicates were used for each experimental condition, * P ≤ 0.05.
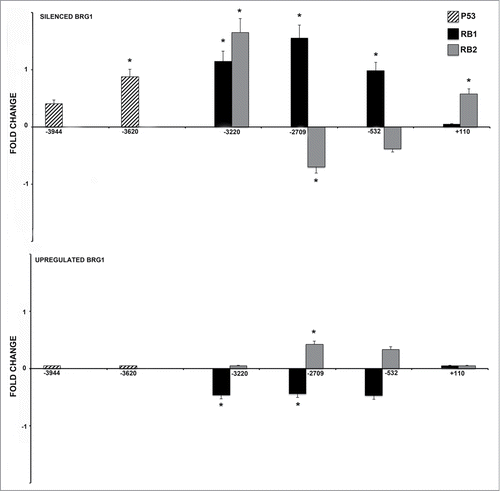
Figure 3. CHIP analysis of the DNMT1, HDAC1 and acetylated H3 binding on the NANOG promoter. MSC with silenced or overexpressed BRG1 was used for CHIP analysis. Panel A shows DNMT1 binding on the E2F site in control cultures and in cells with silenced and over-expressed BRG1. The numbers on the X-axis refer to the nucleotide positions of the identified binding sites. The horizontal bar represents the CpG regions we analyzed with MS-PCR. Panel B shows the HDAC1 binding, and Panel C shows the acetylated H3 binding (H3Ac) on the NANOG promoter in the same experimental conditions reported in A. Real Time PCR data are expressed as arbitrary units. Three biological replicates were used for each experimental condition, * P ≤ 0.05. Y-axis represent cell total DNA bound/input sample according the percent input procedure described in methods.
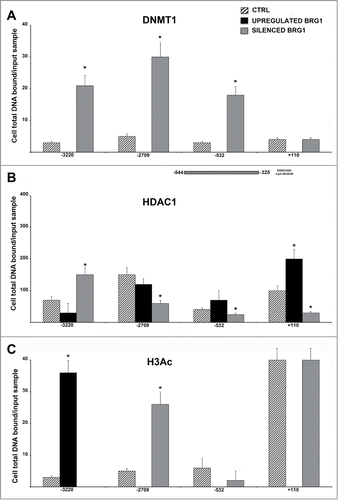
Figure 4. CHIP analysis of BRG1 and BRM binding on the NANOG promoter along with CHART PCR. MSC with silenced or overexpressed BRG1 was used for CHIP analysis. Panel A shows BRG1 binding on the E2F site in control cultures and in cells with silenced and overexpressed BRG1. The numbers on the X-axis refer to nucleotide positions of the identified binding sites. Panel B shows the BRM binding on the NANOG promoter in the same experimental conditions reported in A. Real-time PCR data are expressed as arbitrary units. Three biological replicates were used for each experimental condition, * P ≤ 0.05. Y axis represent cell total DNA bound/input sample according the percent input procedure described in methods. Panel C shows the CHART-PCR analysis to identify heterochromatic regions in the NANOG promoter. This assay evaluates chromatin accessibility by real-time PCR. The more the chromatin is compact, the less it is amenable to digestion by the micrococcal nuclease (MNase). Undigested DNA will provide more copies of target sequences for PCR amplification. We amplified 2 regions (Proximal and Distal) of the NANOG promoter following treatment of DNA with MNase. The upper graph shows results in cells with silenced BRG1. In these cells, the level of amplified DNA, representing heterochromatic DNA, was compared with the control MSC cultures. Changes in the heterochromatic DNA level are expressed as fold changes. The lower graph shows the same analysis in cells with overexpressed BRG1. Three biological replicates were used for each experimental condition, * P ≤ 0.05.
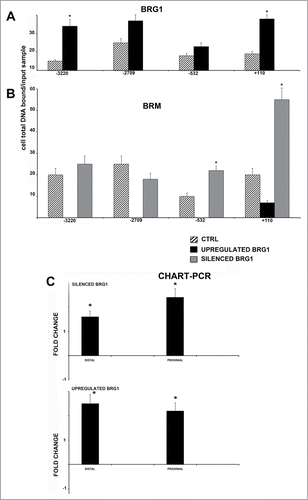
Figure 5. Heat map view representing the functional classification of differentially expressed proteins in upregulated and silenced BRG1, identified by LC-MS/MS analysis. The assessment of enriched biological processes for up- (yellow) and down- (blue) regulated proteins was performed according to gene ontology annotations by using the DAVID software.
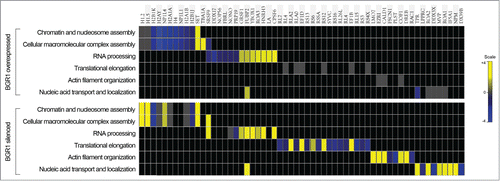
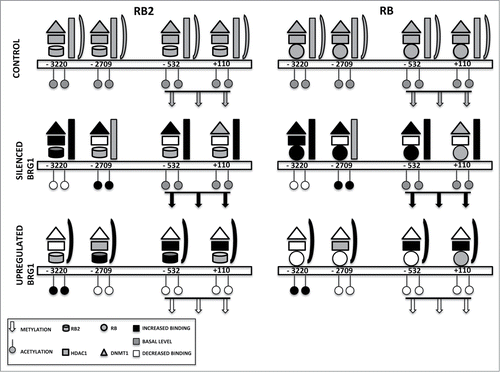
Figure 6. Binding on the NANOG promoter of retinoblastoma proteins and associated factors. The picture depicts the occupancy of 4 E2F binding sites on the NANOG promoter in a basal condition (CONTROL) and in cells with silenced or overexpressed BRG1. For clarity, RB2 binding is reported on the left with the RB1 binding on the right, but the 2 proteins may be present at the same time on the NANOG promoter. According to our model, in cells with silenced BRG1, the inhibition of NANOG transcription may occur mainly through recruitment of DNMT1 through binding with RB1 and/or RB2. This may promote the methylation of CpG dinucleotides in the NANOG promoter. In these cells, the lack of BRG1 is compensated by increased BRM binding. In cells overexpressing BRG1, the DNMT1 and RB1 binding on the NANOG promoter is drastically reduced, while the presence of HDAC1 and RB2 persists on E2F sites. In this condition, the inhibition of NANOG transcription may occur mainly through histone deacetylation.