Figures & data
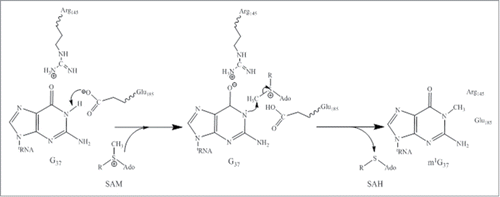
Figure 1. (A) Chemical structure of wybutosine (yW), all appended groups are colored. (B) Secondary structure of yeast tRNAPhe. (C) Chemical structure of SAM. (D) Biosynthetic pathway of yW. Trm5 methylates G37 to produce m1G37; TYW1, a Radical-SAM enzyme, catalyzes the formation of 4-demethylwyosine (imG-14) using pyruvate as a co-substrate; TYW2 appends the α-amino-α-carboxypropyl moiety of SAM to produce 7-aminocarboxypropyl-demethylwyosine (yW-72); TYW3 methylates N4 of yW-72 to produce 7-aminocarboxypropyl-wyosine (yW-58); and TYW4 both transfers a carboxymethyl group and methylates yW-58 to yield yW.
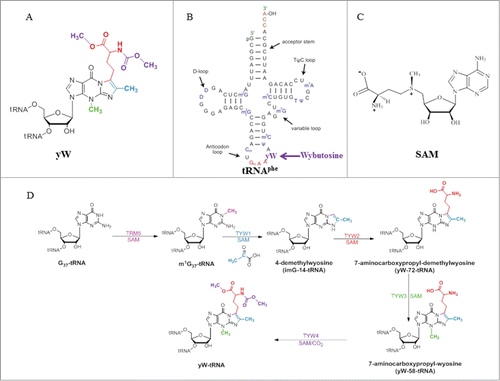
Figure 2. Crystal structure of archaeal Trm5. (A) Structure of Trm5 in complex with SAM (PDB id : 2YX1). The three domains of Trm5 are color coded and labeled. SAM is depicted as a stick model with black for carbon atoms. (B) Ternary structure of Trm5 in complex with tRNALeu and SAM (PDB id : 2ZZM). G37 of tRNALeu is shown as a stick model with orange for carbon atoms. (C) The active site of Trm5 in which the side chains of R145 and the proposed catalytic residue E185 are shown as stick models. are made using PyMOL (www. pymol. org).
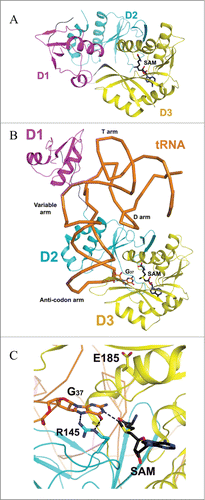
Figure 4. Crystal structure of yeast TYW4. (A) Apo structure of TYW4 (PDB id : 2ZWA). The three domains of TYW4 are colored and labeled. The N-terminal helices are shown as yellow. (B) Structure of TYW4 in complex with SAM (PDB id : 2ZW9). SAM is depicted as a stick model with black for carbon atoms. The N-terminal helix is shown as yellow. (C) The active site of TYW4 in which the side chains of Y229 and R88, and SAM are shown as stick models. The flexible loop of C-domain is shown in magenta and is labeled. The substrate binding site is labeled.
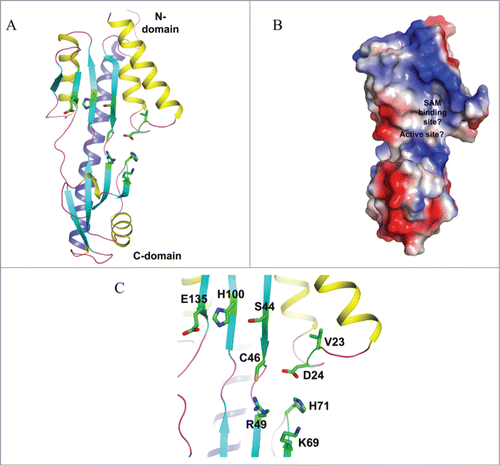
Figure 5. Crystal structure of a putative TYW3 from Archaeoglobus fulgidus. (A) Apo structure of TYW3 (PDB id : 2QG3). The secondary structural elements, β-strands, α-helices, and loops, are colored as cyan, yellow, and light-magenta. The C-terminal α-helix is colored as dark blue. The side chains of invariant residues are shown with stick models. (B) Electrostatic surface potential of TYW3. Blue, red, and white patches present the positively, negatively-charged, and neutral regions of the protein surface, respectively. The presumed SAM binding pocket and the active site are labeled. (C) The presumed SAM-binding and the active sites of TYW3. The side chains of strictly conserved residues are shown as stick models.
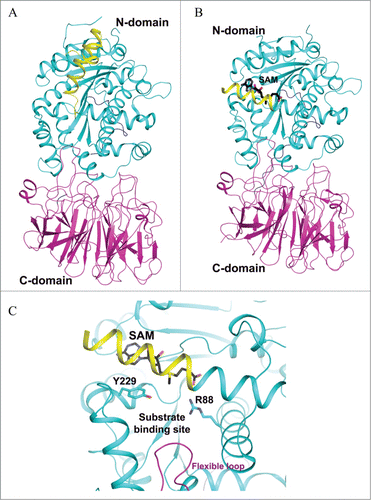
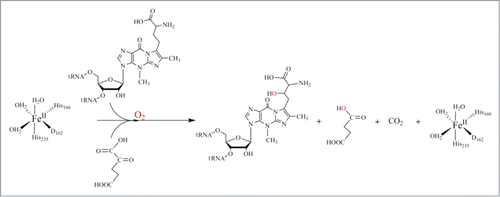
Figure 6. Proposed catalytic reaction mechanism for both methylation (A) and carboxymethylation (B) for TYW4.
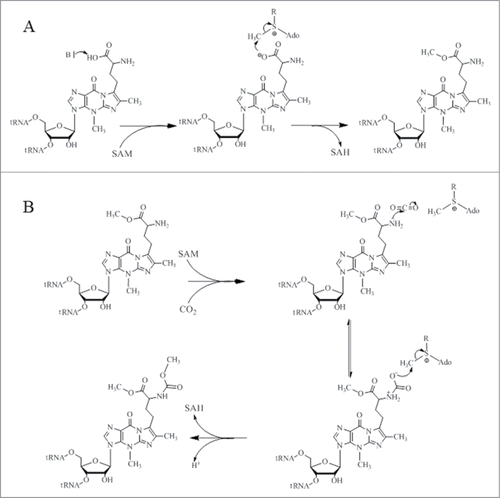
Figure 7. Crystal structure of P. horikoshii TYW2. (A) SAM-complexed structure of TYW2 (PDB id : 3A25). The two domains of TYW2 are color coded and labeled. SAM and the substrate binding pocket are labeled. (B) The active site of TYW2 in which the side chains of M107, N112, R116, N112, and SAM are shown as stick models. SAM and the location of the substrate binding pocket are labeled.
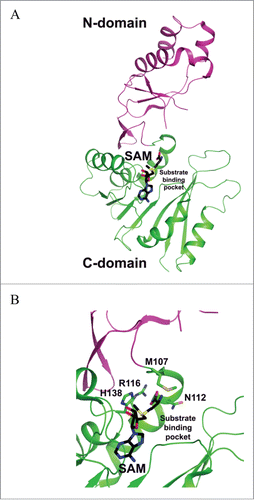
Figure 8. Crystal structures of TYW1. (A) Apo structure of archaeal TYW1 (PDB id : 2Z2U). The side chains of invariant cysteine residues are depicted as stick models and are labeled. (B) Apo structure of P. horikoshii TYW1 (PDB id : 2YX0). The side chains of invariant cysteine residues are depicted as stick models and are labeled.
Figure 9. Proposed catalytic reaction mechanism of holo-TYW1. Radical-SAM cluster I and cluster II are sketched as cubes, non-cysteinyl irons are shown in blue (cluster I) and red (cluster II), (A): SAM and pyruvate binding to cluster I and II respectively (reaction 1). Reduction of cluster I (reaction 2). Reductive cleavage of SAM in presence of m1G37-tRNA.
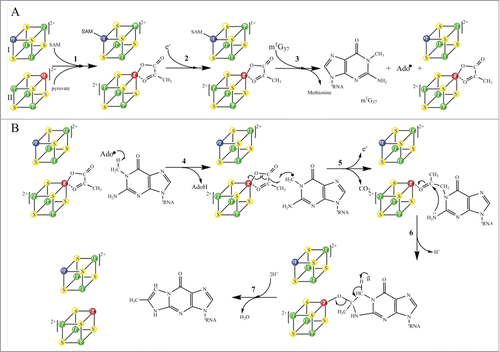
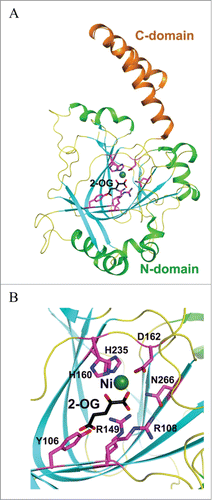
Figure 11. Crystal structures of human TYW5. (A) Structure of hTYW5 in complex with Ni2+ and 2-oxoglutarate (PDB id : 3AL6). The two domains are depicted with different colors. Ni ion is shown as a dark green sphere, whereas 2-oxoglutarate (2-OG) and side chains of residues in the active site are shown with stick models. (B) The active site of TYW5. The side chains of invariant residues in the active site, Ni ion, and 2-OG are depicted as stick models and are labeled.