Figures & data
Table 1. Minimal, average and maximal percentage of modified residues per tRNA molecule in 7 groups of organisms analyzed in this work
Figure 1. Consensus tRNA secondary structure presented in the “cloverleaf” form with the universal numbering system.Citation26
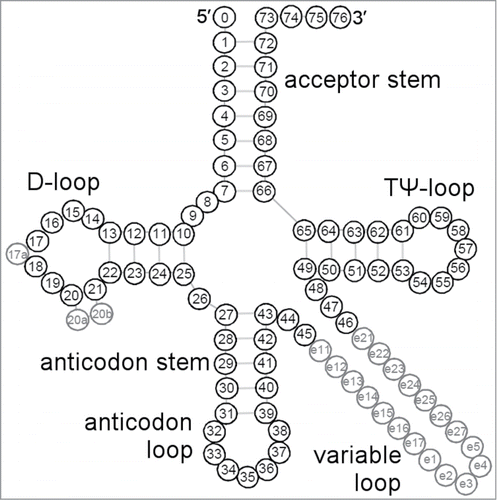
Figure 2. (A) Distribution of modified residues in tRNA sequences from E. coli as presented by the tRNAmodviz server. Percent of modified residues within each position is mapped onto the consensus tRNA secondary structure (small black and white pie charts within the cloverleaf). (B) Frequency of A, U, C or G bases occurrence and their derivatives in tRNA sequences from E. coli. (C) Color pie charts which present detailed information for chosen position in tRNA sorted by the originating base, sorted by the presence of modification or covering modifications only – these options the user can choose when working with tRNAmodviz.
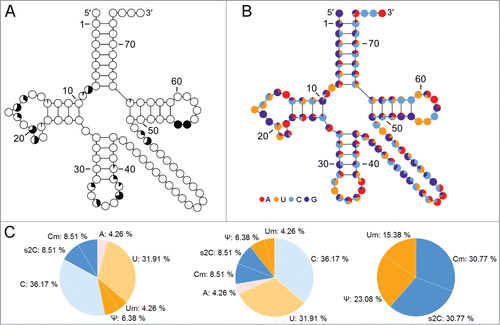
Figure 3. Modification profile for tRNA sequences from Gram-negative bacteria (62 sequences from 8 species). The pie charts within each position in the cloverleaf correspond to the percentage of all modified nucleosides (modified being drawn in black). In the tables the series of numbers next to the series of symbols indicate the frequency of occurrence of listed nucleosides at the particular position. The last number corresponds to the frequency of occurrence of the encoded unmodified nucleoside. The numbering of the residues is presented in . The list of species from which the analyzed tRNA sequences originate is provided in Supplementary Table.1.
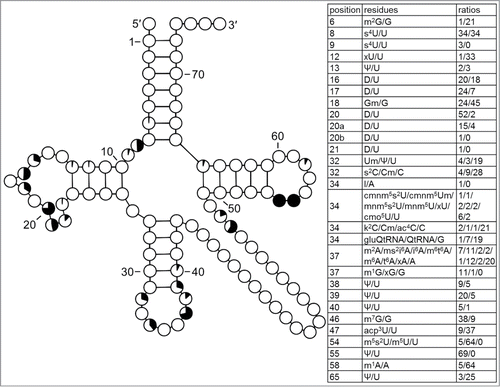
Figure 4. Modification profile for tRNA sequences from Gram-positive bacteria (72 sequences from 9 species). For the description of the tables and cloverleaf content see the legend for . The numbering of the residues is presented in . Among sequences considered for this group there are 2 tRNAGly isoacceptors from Staphylococcus epidermidis which probably do not function in protein synthesis.Citation59 These 2 lack the conserved pseudouridine in position 55. The list of species from which the analyzed tRNA sequences originate is provided in Supplementary Table.1.
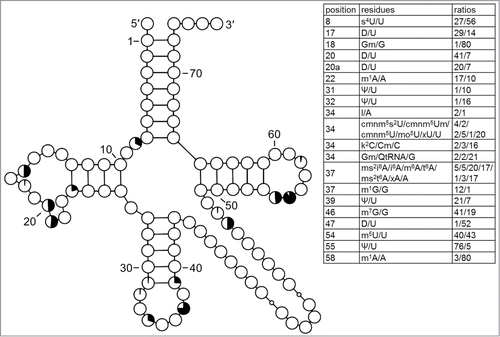
Figure 5. Modification profile for tRNA sequences from cytosol of eukaryotic single cell organisms, Fungi and Metazoa (173 sequences from 22 species). For the description of the tables and cloverleaf content see the legend for . The numbering of the residues is presented in . According to the original work xG37 in 2 R. norvegicus tRNALeu was converted to m1G by treatment with alkali.Citation60 The list of species from which the analyzed tRNA sequences originate is provided in Supplementary Table.1.
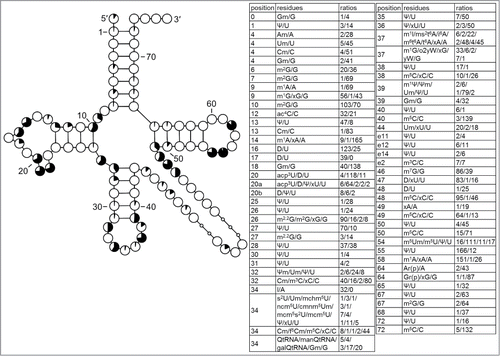
Figure 6. Modification profile for tRNA sequences from cytosol of Viridiplantae (55 sequences from 14 species). For the description of the tables and cloverleaf content see the legend for . The numbering of the residues is presented in . xG64 in Scenedesmus obliquus and Triticum aestivum is most probably Gr(p), like in cytoplasmic tRNAIni from S. cerevisiae. The list of species from which the analyzed tRNA sequences originate is provided in Supplementary Table.1.
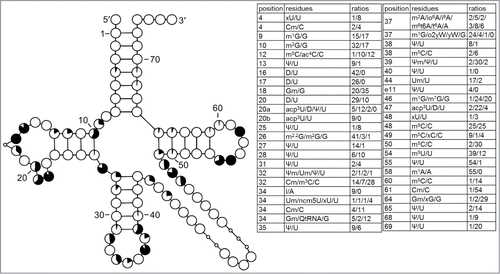
Figure 7. Modification profile for tRNA sequences from mitochondria (124 sequences from 18 species). For the description of the tables and cloverleaf content see the legend for . The numbering of the residues is presented in . Note that sequences from Ascaris suum have a lot of deletions and as a result their alignment is problematic. In the majority of cases xA37 is i6A37 or ms2i6A. In Neurospora crassa tRNATyr m5C is present in position e21 or 48.Citation61 In this compilation m5C in e21 and xC48 were kept. The list of species from which the analyzed tRNA sequences originate is provided in Supplementary Table.1.
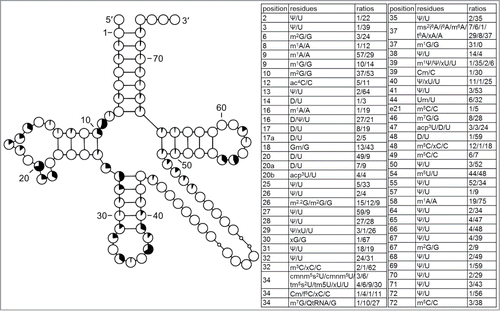
Figure 8. Modification profile for tRNA sequences from plastids (38 sequences from 12 species). For the description of the tables and cloverleaf content see the legend for . The numbering of the residues is presented in . The list of species from which the analyzed tRNA sequences originate is provided in Supplementary Table.1.
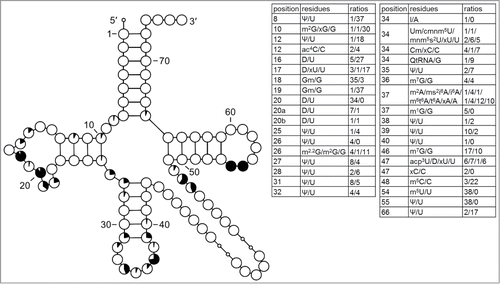
Figure 9. Modification profile for tRNA sequences from Euryarchaeota (60 sequences, mostly Haloferax volcanii). For the description of the tables and cloverleaf content see the legend for . The numbering of the residues is presented in . xA57 present in initiator tRNA from Thermoplasma acidophilum is not m1I.Citation27 The list of species, from which the analyzed tRNA sequences originate, is provided in Supplementary Table.1.
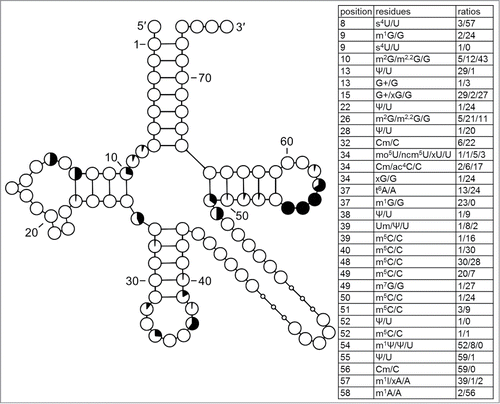
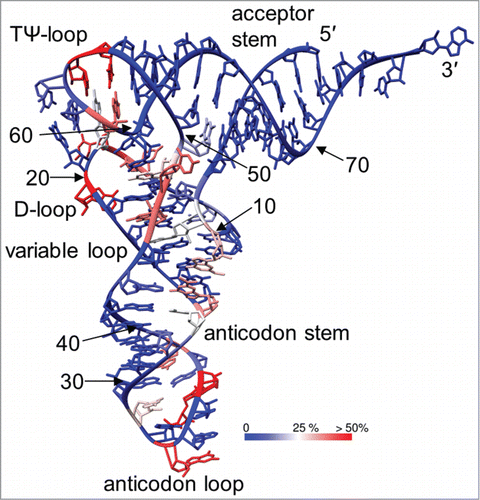
Figure 10. Frequency of modification marked on the structure of yeast tRNAPhe (PDB ID: 1ehz). The color scale from blue via white to red indicates the percentage in which each position is modified when all sequenced tRNAs are considered. The arrow indicates the interface of interactions between the D- and TΨ-loops.