Figures & data
Figure 1. tRNA modification defects and phenotypes in higher eukaryotes. Schematic representation of a tRNA. Modified nucleosides that have been linked to phenotypes in higher eukaryotes are indicated as red circles. The color inside the circle denotes the type of defect observed. Chemical modifications and their causative genes (in brackets) are linked to the respective nucleoside. Gray or black residues depict nucleosides that are either unmodified or not linked to phenotypes. Abbreviations of the nucleosides follow the nomenclature of Modomics (http://modomics.genesilico.pl/).
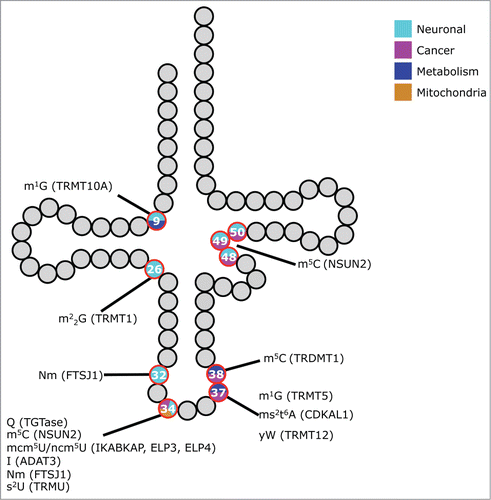
Figure 2. RNA modification genes associated with human malignancies. Schematic representation of various cancers for which increase (red) or decrease (blue) of tRNA modification gene copy number, or expression level, has been reported. (Source: Cosmic and Oncomine).
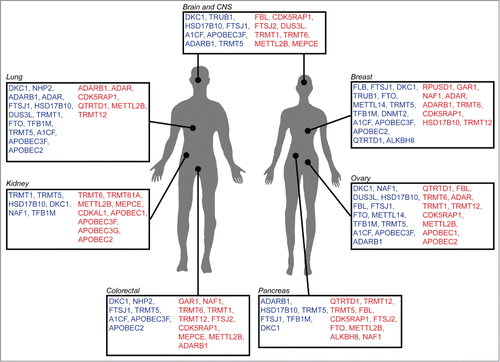
Table 4. Factors to consider:
Figure 3. Models of how RNA modification defects cause phenotypes. Comparison of different scenarios in an unperturbed (left) and pathogenic (right) situation. (A) A metabolic pathway is blocked, leading to the absence of modified RNA (indicated by an asterisk) and the build up of a different metabolite (in this case Z). (B) Ribosomes (orange) translating an mRNA, which contains a region that is difficult to translate (red box). In the pathogenic situation translation is perturbed, leading to a lower amount of protein. (C) As in (B). Perturbed translation prevents the folding of some proteins into their native state resulting in perturbed protein homeostasis. (D) tRNA fragments cause a slowdown of translation.
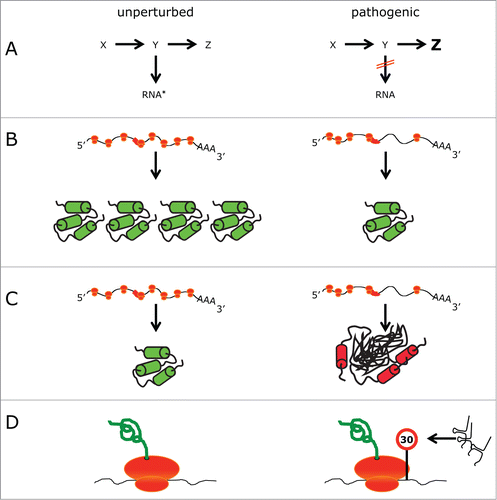
Table 1. RNA modification genes associated with disease in humans
Table 2. Mouse models for RNA modification genes.
Table 3. Drosophila and zebrafish models for RNA modification genes.
