Figures & data
Figure 1. GC% distribution and DNA replication timing of human chromosome 21q in the THP-1 cell line, and in NPCs and ESCs. Replication timing in chromosome 21q in THP-1 cells (data from Watanabe et al. 2002), NPCs and ESCs. The latter data were obtained by analysis of information held on ReplicationDomain (http://www.replicationdomain.com). The “y” axis on each graph indicates the estimated numerical value for DNA replication timing obtained from ReplicationDomain. The value “0” indicates medium replication timing. The upper horizontal line and the bottom horizontal line of each graph indicate the value of 2.4 (very early replication timing) and -2.4 (very late replication timing), respectively. The “x” axis indicates genomic position of the gene. The positions of 3 genes (APP, GRIK1 and DSCAM) are indicated by the green dotted lines. The bottom panel shows GC% distribution along chromosome 21q.
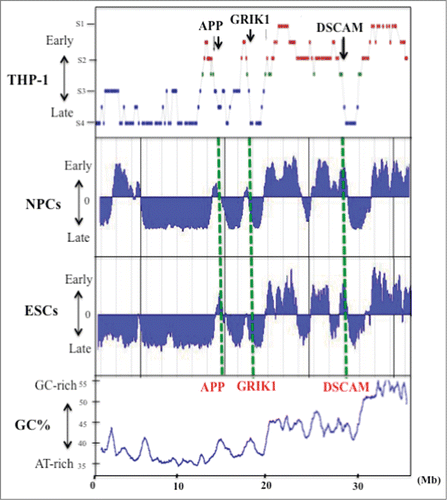
Figure 2. DNA replication timing for genomic regions in and around the large genes APP and GRIK1 (A) and DSCAM (B) in THP-1, Jurkat and HCT116 cells, and in NPCs, ESCs and lymphoblastoid cells. Replication timing in THP-1, Jurkat and HCT116 cells were determined as described in the “Materials and Methods.” The “x” axis of each graph indicates the genomic position of the gene as given by the Ensembl Genome Brower (www.ensembl.org/). The position and the range of each gene are indicated by the red arrows. The PCR primer sets used in the present study and the replication timings are listed in Supplemental Table 1. Replication timings in NPCs, ESCs and lymphoblastoid cells were obtained by analysis of information held on ReplicationDomain (http://www.replication-domain.com). The “y” axis on each graph indicates the estimated numerical value for DNA replication timing obtained from ReplicationDomain. The value “0” indicates medium replication timing. The upper horizontal line and the bottom horizontal line of each graph indicate the value of 2.4 (very early replication timing) and -2.4 (very late replication timing), respectively. The “x” axis of each graph indicates the genomic position of the gene, which was obtained from ReplicationDomain. The position and range of each gene in NPCs, ESCs and lymphoblastoid cells are indicated by the dotted red lines. “Lymph.” indicates lymphoblastoid cells.
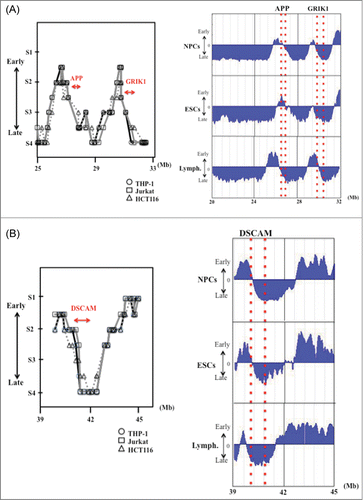
Table 1. DNA replication timing in NPCs, ESCs and lymphoblastoid cells and lengths of human neuronal glutamate receptor genes
Figure 3. DNA replication timings of selected neuronal glutamate receptor genes in NPCs, ESCs and lymphoblastoid cells. The position or the range (large genes) of each gene is indicated by a red dotted line or 2 dotted lines for large genes. (A) Replication timing of AMPA glutamate receptor family (GRIA1–4). (B) Examples of replication timing of other large (> 200 kb) glutamate receptor genes. The “y” axis on each graph indicates the estimated numerical value for DNA replication timing obtained from ReplicationDomain. Average replication timing data for each cell line is shown. The value “0” indicates medium replication timing. The upper horizontal line and the bottom horizontal line of each graph indicate the value of 2.4 (very early replication timing) and -2.4 (very late replication timing), respectively. “Lymph.” indicates lymphoblastoid cells.
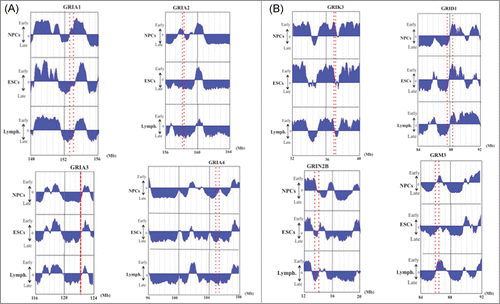
Table 2. DNA methylation of 5′-upstream genomic regions of GRIK1, GRIA2 and GRIA4 genes in NPCs and ESC
Figure 4. Examples of the precise replication timing patterns of large genes (>200 kb). (A) APP (260kb), (B) GRIA1 (324 kb) and (C) GRIN2B (418 kb). Replication timing profiles in 6 cell lines (NPCs and ESCs, lymphoblastoid, myoblast and fetal lung fibroblast) were obtained from the online database ReplicationDomain. The “y” axis on each graph indicates the estimated numerical value for DNA replication timing obtained from ReplicationDomain. The value “0” indicates medium replication timing. The upper horizontal line and the bottom horizontal line of each graph indicate the value of 2.4 (very early replication timing) and -2.4 (very late replication timing), respectively. The direction of transcription (5′ to 3′) in the genes is indicated by the red arrow. “Lymph.,” “Myo.” and “Fibro.” indicate lymphoblasts, myoblasts and fetal lung fibroblasts, respectively.
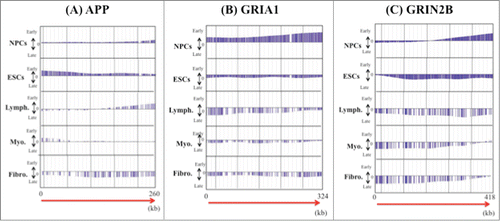
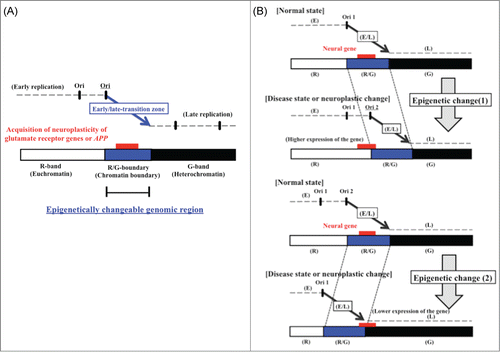
Figure 5. (A) Model of high-risk/high-return regions of the human genome located in transition regions of replication timing as the epigenetic molecular basis of neuroplastic changes. The model proposes that modifications of the chromatin structure during development of neural diseases (or neuroplastic changes) affect the timing of firing of replication origins. (B) Postulated changes in replication timing of a neural disease gene located in a transition zone. The panels illustrate how replication timing might switch from mid S phase to early or late S phase due to an increase (top panel) or decrease (bottom panel) in the number of active early replication origins at the edge of the early replication zone. Additionally, the chromatin environment of the neural disease gene might switch from that of an R/G chromosome band boundary to an R band or to a G band. Stalling of the replication fork in the vicinity of neural disease genes might also induce chromosomal amplifications (such as triplet repeat expansions) or chromosome rearrangements that affect gene function, possibly through influencing the rate of expression. The position of the neural disease gene (large gene) is indicated by the red rectangle. E, early replication zone; L, late replication zone; E/L, early/late-switch region; R, R band; G, G band; R/G, R/G band boundary; Ori, replication origin.